Introduction
The population of overweight people is increasing globally every year, and the number of people with diabetes is also rapidly increasing (WHO, 2017). The excessive intake of calories can lead to many chronic diseases such as obesity, type 2 diabetes mellitus, hyperlipidemia, certain cancers, and cardiovascular diseases (Etoundi et al., 2010). Especially, obesity is the most common complication of hyperlipidemia and type 2 diabetes mellitus, and they share common causative factors, chemical abnormalities, and clinical complications (Poothullil, 1993; Shamseddeen et al., 2011). Several studies reported that obesity and weight gain are associated with an increased risk of diabetes (Ford et al., 1997; Resnick et al., 2000).
Lactic acid bacteria (LAB) are common microorganisms in foods and are also present in the natural intestinal microbiota of humans and most animals (Tannock, 1995). LAB is frequently used as probiotics and for the fermentation of food products. Probiotics are defined as live microorganisms that, when administered in adequate amounts, provide a health benefit to the host (Joint FAO/WHO, 2002). The criteria for the selection of probiotic bacteria for food applications as starters should be carefully checked for each particular strain. When selecting probiotic starters, several aspects such as technological and functional properties and safety of the strains have to be taken into consideration (Ahmadova et al., 2013). The criteria for the selection of probiotics include antibiotic susceptibility, prevention of microbial pathogenicity, tolerance to gastrointestinal conditions (bile and acid), biogenic amine production, and ability to adhere to the gastrointestinal epithelium (Collins et al., 1998; Ouwehand et al., 2002; Rönkä et al., 2003).
In this study, we investigated the anti-obesity effect of Lactibacillus plantarum K10, which was selected from among LAB isolated from kimchi, and studied the physiological characteristics to determine its potential as starter for functional food products.
Materials and Methods
The strain K10 was isolated from the homemade kimchi in a modified MRS medium (Lim et al., 2011). The strain was incubated in Man, Rogosa, and Sharpe broth (MRS, Difco, USA) as growth medium at 37°C for 18 h.
Bacterial genomic DNA samples were extracted using the InstaGeneTM Matrix (Bio-Rad, USA). The primers 27F 5’ (AGA GTT TGA TCM TGG CTC AG) 3’ and 1492R 5’ (TAC GGY TAC CTT GTT ACG ACT T) 3’ were used for PCR. PCR reaction was performed with 20 ng of genomic DNA as template in a 30 µL reaction mixture by using EF-Taq (Solgent, Korea) as follows: activation of Taq polymerase at 95°C for 2 min, 35 cycles of 95°C for 1 min, 55°C, and 72°C for 1 min each was performed, finishing with a 10-min step at 72°C. The amplification products were purified with a multiscreen filter plate (Millipore Corp., USA). Sequencing reaction was performed using a PRISM BigDye Terminator v3.1 Cycle Sequencing Kit (Applied Biosystems, USA). DNA samples containing the extension products were added to Hi-Di formamide (Applied Biosystems). The mixture was incubated at 95°C for 5 min followed by 5 min on ice and then analyzed by ABI Prism 3730XL DNA analyzer (Applied Biosystems).
A modified version of the method of α-amylase activity determination by Kusano et al. (2011) was used. Porcine pancreas α-amylase was purchased from Sigma (USA). The substrate was prepared by boiling 0.5% soluble starch in distilled water for 5 min, and then cooling to room temperature. The sample (100 µL) and substrate (500 µL) were mixed in 400 µL of 0.04 M phosphate buffer (pH 5.8). After that, 0.5 mg/mL α-amylase solution (100 µL) was added, and the solution was incubated at 25°C for 10 min. The reaction was stopped by adding 100 µL 0.1M HCl, and then 100 µL of the solution was reacted with 1.5 mL iodine solution for 30 min at room temperature. The absorbance was measured at 660 nm by a microplate reader (Spectramax Plus 384, Molecular Devices Corp., USA).
α-glucosidase inhibition assay was performed as previously described (Kim et al., 2011) but modified as follows. Inhibitory activity was measured using α-glucosidase from Saccharomyces cerevisiae (Sigma, USA). α-glucosidase (50 µL, 0.75 U/mL) and 0.2 M potassium phosphate buffer (pH 6.5, 50 µL) were mixed with 50 µL of test sample. After pre-incubation at 37°C for 15 min, 3 mM p-nitrophenol-αD-glucopyranoside (pNPG, 100 µL) was added. Enzymatic reaction was allowed to proceed at 37°C for 10 min and was stopped by the addition of 750 µL of 0.1 M Na2CO3. 4-Nitrophenol absorption was measured at 405 nm using a microplate reader.
The method of Lee et al. (1993) for lipase activity determination was modified. Pancreatic lipase activity was measured using porcine pancreatic lipase (Sigma). 0.1 mg/mL of a sample solution dissolved in water and 0.167 mM p-Nitrophenylpalmitate (PNP) solution and 0.061 M (pH 8.5) Tris-HCl buffer were mixed in the well of a plate, and 0.3 mg/mL of the lipase solution was then added to start the enzymatic reaction. After incubation at 25°C for 10 min, its absorbance was measured at 405 nm using a microplate reader.
3T3-L1 cells were obtained from the American Type Culture Collection (ATCC, USA) and were cultured at 37°C in a humidified 5% CO2 atmosphere. 3T3-L1 cells were cultured as described by Hemati et al. (1997). To describe the method briefly, the cells were grown in Dulbecco’s modified Eagle’s medium (DMEM, Gibco, USA) containing high glucose supplemented with 10% bovine calf serum (BCS, Gibco) and 1% penicillin/streptomycin in six well culture plates. Two days later, confluence cells were cultured in an adipocyte differentiation cocktail medium containing 0.5 mM 3-isobutyl-1-methylxanthine (IBMX, Sigma), 1 µM dexamethasone (Dex, Sigma), and 5 μg/mL insulin (Sigma) in DMEM supplemented with 10% fetal bovine serum (FBS, GIBCO) for 2 days. The differentiation was complete after 6 days.
L. plantarum K10 was incubated at 37°C for 18 h in MRS broth. After culturing L. plantarum K10, all of the strains were harvested in a refrigerated centrifuge (1,500×g for 15 min at 4°C) and washed three times with distilled water to remove any remaining MRS broth. The washed L. plantarum K10 was freeze-dried and re-suspended in distilled water at a concentration of 10 mg/mL and homogenized for 50 s followed by 1 min of rest (repeated 3 times) using a sonicator. The 3T3-L1 cells were treated with 100 µg/mL of L. plantarum K10 (109 CFU/mL).
Intracellular lipid accumulation was measured using oil red O (Sigma). Oil red O staining of 3T3-L1 cells was done using a modified version of the method described by Ramirez-Zacarias et al. (1992). 3T3-L1 cells were washed with PBS (phosphate-buffered saline) twice, fixed with 10% formaldehyde/PBS at 4°C for 1 h, and stained with filtered oil red O solution (stock solution: 3.5 mg/mL in isopropanol; working solution: 60% oil red O stock solution and 40% distilled water) at room temperature for 30 min. The quantification of lipid accumulation was achieved by oil red O obtained from stained cells with isopropyl alcohol and measured spectro-photometrically at 520 nm. The material stained with oil red O was expressed on a per cell basis using the cell number determined from similar plates. The percentage of the material stained with oil red O relative to the control wells containing the cell culture medium without compounds was calculated as A520 nm (Q180)/A520 nm (control)×100.
The antibiotic susceptibility of L. plantarum K10 was tested by the broth microdilution procedure (Phillips et al., 1991) with the following modifications. LAB Susceptibility test Medium with cysteine (LSM-C) was used as medium. Essentially, LSM-C consists of a mixture of Iso-Sensitest broth (90%) and MRS broth (10%), supplemented with 0.3 g/L L-cysteine (Klare et al., 2007). L. plantarum K10 was grown at 37°C for 18 h in MRS broth and inoculated into LSM-C medium (final concentration of 105–106 CFU/mL) supplemented with antibiotics (amikacin, gentamicin, kanamycin, streptomycin, ampicillin, penicillin-G, oxacillin, bacitracin, ciprofloxacin, tetracycline, clindamycin, erythromycin, rifampicin, vancomycin, and chloramphenicol, Sigma; polymyxin B, Difco) at various concentrations in a two-fold dilution step. The minimal inhibitory concentration (MIC) coincides with the lowest antibiotic concentration that prevents cell growth after incubation at 37°C for 18–20 h. All tests were performed in triplicate.
An API ZYM kit (bioMerieux, France) was used to study enzyme activity. L. plantarum K10 was grown at 37°C for 18 h in MRS broth. Sediment from the centrifuged broth culture was used to prepare the suspension at 105–106 CFU/mL. After inoculation, the cultures were incubated for 5 h at 37°C. The addition of a surface active agent (ZYM A reagent) in the cupules facilitated the solubilization of the ZYM B reagent in the medium. Color was allowed to develop for at least 5 min, and values ranging from 0 to 5 (corresponding to the colors developed) were assigned. The approximate number for the free nmol hydrolyzed substrate was determined based on the color strength: 0, negative reaction; 1, 5 nmol; 2, 10 nmol; 3, 20 nmol; 4, 30 nmol; 5, 40 nmol or higher.
L. plantarum K10 grown in MRS broth at 37°C for 18 h were cultured in a special medium according to Chang and Chang’s method (2012) for 24 h at 37°C. LB agar (pH 5.0; Difco) containing 0.25% glycerol, 0.006% bromocresol purple, and 0.1% precursor amino acid (tyrosine, histidine, ornithine, and lysine, respectively; Sigma) were used. The strain was washed three times with distilled water, and 10 µL of sample was loaded onto sterile filter paper discs (8 mm diameter, Advantec, Japan). Biogenic amine production was determined by the color change of the pH indicator bromocresol purple in the medium.
pH tolerance was tested as described by Clark et al. (1993). Solutions of 37% HCl in double-distilled water were adjusted to pH levels of 2.0, 3.0, and 4.0. Sterile double-distilled water (pH 6.4) served as the control. 10 mL of each pH solution was transferred into sterile test tubes. 1 mL of stock culture containing approximately 109 CFU/mL of L. plantarum K10 using MRS agar containing 0.05% cysteine was then transferred into each of the four pH solutions. The pH solutions containing L. plantarum K10 were then incubated anaerobically at 37°C, followed by intermittent plating after 1, 2, and 3 h to stimulate the survival of L. plantarum K10 under pH conditions common to the human stomach. Samples from the pH solution were re-suspended and subjected to serial dilutions. About 100 μL of the abovementioned sample solution was spread onto the surface of the BCP (bromo cresol-purple) plate count agar plates and incubated anaerobically at 37°C for 48 h.
Bile tolerance was tested as described by Gilliland and Walker (1990). L. plantarum K10 was grown at 37°C for 18 h in MRS broth. Each 1% of the L. plantarum K10 strain culture was inoculated into sterilized MRS broth containing 0.05% L-cysteine (Sigma) with or without 0.3% oxgall (Sigma), and then the growth potential was compared in the presence of the bile. Afterward, the cultures were incubated anaerobically at 1 h intervals for 7 h at 37°C. All pour plates were incubated anaerobically at 37°C for 48 h using BCP plate count agar (Eiken Chemical Co., Japan).
Antimicrobial activity was tested as described by Gilliland and Speck (1977). Escherichia coli ATCC 21985, Salmonella Typhimurium ATCC 14028, Listeria monocytogenes ATCC 15313, and Staphylococcus aureus ATCC 6538 were obtained from the Korean Culture Center of Microorganisms (KCCM, Korea). E. coli were enumerated on Eosin Methylene Blue agar (EMB, Difco), S. Typhimurium, on Bismuth sulfite agar (Difco), L. monocytogenes, on Oxford medium (LAB122, supplement X122; Lab M, Lancashire, UK), and Staphylococcus aureus, on Baird Parker agar (Difco). All the plates were incubated for 48 h at 37°C. Both control culture and associative culture were incubated for 6 h at 37°C. At the end of the incubation period, the samples were removed and placed in an ice bath pending analysis. The number of CFU of pathogens per mL was determined using the appropriate selective medium. Percentages of inhibition were determined using the following formula:
The adhesion of L. plantarum K10 was studied using the HT-29 intestinal epithelial cell line (Kim et al., 2008). HT-29 cells were obtained from the Korea Cell Line Bank (Korea). The cells were cultured at 37°C in a 5% CO2-95% air atmosphere in RPMI 1640 (Gibco) supplemented with 10% FBS. The sub-cultured (3 times) L. plantarum K10 was harvested by centrifugation at 16,000×g rpm for 3 min, and then washed three times with PBS to remove any remaining MRS broth. The washed bacteria were then re-suspended in an RPMI 1640 medium to an optical density at 600 nm (OD600) of 0.5 (approximately 107 CFU/mL). The re-suspended bacteria were appropriately diluted and plated on a BCP plate count agar. To investigate the adhesion activity, post-confluent HT-29 cells were washed twice with PBS. After washing, 1 mL of the bacteria in the RPMI 1640 medium was added to each well of the tissue-culture plate (12 wells), which was then incubated for 2 h. After incubation, the cells were washed five times with sterile PBS and harvested with a trypsin-EDTA (0.25% trypsin and 0.02% EDTA, Gibco). It was appropriately diluted and plated on a BCP plate count agar to determine the number of viable cell-associated bacteria.
The Committee on the Ethics of Animal Experiments of Handong Global University authorized and permitted the animal experiment. Five-week-old C57BL/6J male mice were arranged by Koatec (Korea) and were housed in a controlled environment with free access to filtered water and food. The controlled environment was set to 23±1°C and 55±10% humidity in a 12 h light/dark cycle. For the first week of acclimation, the mice had an identical diet of normal chow; after the acclimation, the mice were randomly assigned into groups of 6 with different diets for 12 wk. The high-fat diet (HFD) consists of 40% carbohydrates, 45% fat, and 15% protein. Probiotic strains were freeze-dried and incorporated with 3 grams of HFD diet with 109 CFU/mice/day. The weight of the mice and their feed consumption were measured once a week. At the end of this experiment, the mice were anesthetized with diethyl ether inhalation and samples were collected, with their weights measured. All the samples were stored under –80°C without repeated freeze-and-thaw steps.
Intestinal permeability was determined by analyzing the FITC-dextran concentration with a fluorescence spectro-photometer at excitation wavelength of 485 nm and emission wavelength of 535 nm. This experiment follows the protocol listed in the Bio-protocol (Gupta and Nebreda, 2014). Briefly, mice were fasted for 4 h before administering FITC-dextran (Sigma) to each mouse (44 mg/100 g body weight) by oral gavage. After 4 h, mice were anesthetized with cervical dislocation, with the blood collected by cardiac puncture. The blood was centrifuged at 4°C, 12,000×g for 3 min, and serum was then collected. The serum was diluted in an equal volume of PBS (pH 7.4), and the concentration of FITC-dextran was analyzed with a fluorescence spectrophotometer (HTS-7000 Plus-plate-reader, PerkinElmer, USA). Standard curves were obtained by serially diluting FITC-dextran (0, 125, 250, 500, 1,000, 2,000, 4,000, 6,000, 8,000 ng/mL) with untreated plasma with PBS (1:2 [vol/vol]).
Each experiment was carried out in triplicate, and the results were expressed as mean±standard deviation (SD). Statistical analysis was performed using a statistical analysis system (SAS, SAS Institute, Inc., USA). The significance of the differences was analyzed by conducting a one-way analysis of variance (ANOVA) with Duncan’s multiple range tests. Values of p<0.05 were considered statistically significant.
Results and Discussion
After PCR amplification using universal primers targeting 16S rDNA as well as the following sequence analysis, it was identified as L. plantarum with similarity of 100% (data not shown). Based on the results of previous studies, it was named L. plantarum K10.
To select strong inhibitory activities of α-amylase, α-glucosidase, and pancreatic lipase, we determined the α-amylase, α-glucosidase, and pancreatic lipase inhibitory activities of 167 kinds of isolated strain in Kimchi. After being incubated in MRS broth at 37°C for 18 h, the L. plantarum K10 strain exhibited α-amylase, α-glucosidase, and pancreatic lipase inhibitory activity of 94.66±4.88%, 99.78±0.28%, and 87.40±1.41%, respectively, compared with the control (Table 1). Both natural and synthetic pancreatic lipase inhibitors are effective in preventing obesity because they inhibited intestinal lipid absorption (Hirose et al., 2013). Because the dietary habits of Korea include much more carbohydrates than those of western countries, the mechanism of inhibiting the absorption of carbohydrates should be combined with the mechanism of inhibiting fat absorption to improve obesity (Jang and Jeong, 2010).
| Strain | α-amylase inhibitory activity (%) | α-glucosidase inhibitory activity (%) | Lipase inhibitory activity (%) |
|---|---|---|---|
| L. plantarum K10 | 94.66±4.88 | 99.78±0.28 | 87.40±1.41 |
Obesity is associated with the degree of differentiation of preadipocytes into adipocytes and enlargement of adipocytes in adipose tissues (Wang and Jones, 2004). The effect of L. plantarum K10 in 3T3-L1 adipocyte stained with oil red O is shown in Fig. 1. A cell treated with K10 showed about 32.61% reduction in lipid accumulation compared with the untreated control (p<0.05, Fig. 1A). As shown in Fig. 1B, L. plantarum K10 exhibited a decrease in lipid accumulation rounded cells compared with the untreated control cells when visualized by staining. The oil red O staining test with L. plantarum K10 revealed an observable decrease in lipid accumulation in 3T3-L1 cell during adipogenesis, suggesting that K10 suppresses adipocyte differentiation. Therefore, it implies that K10 has an anti-obesity effect through inhibition pancreatic lipase and differentiation of 3T3-L1 preadipocytes by suppressing lipid accumulation.
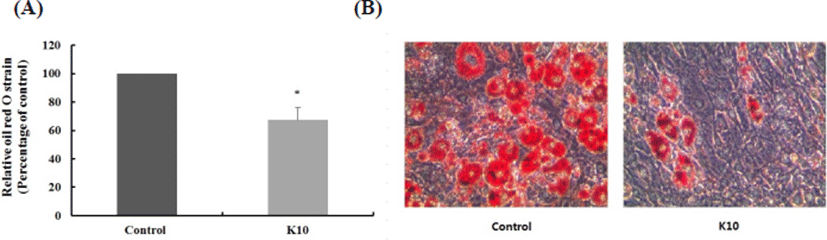
Table 2 shows the MIC values obtained for the 16 kinds of different antibiotics tested in L. plantarum K10. The clindamycin MIC value was lowest among the antibiotics. L. plantarum K10 showed the highest vancomycin MIC. This is because Lactobacillus strains have intrinsic resistance to glycopeptides (Perichon and Courvalin, 2000). The 46 kinds of isolated L. plantarum strains were reported to have an MIC range of 0.125–2 µg/mL in ampicillin (Klare et al, 2007). The MIC of L. plantarum K10 showed high resistance to ampicillin 2,048 µg/mL. Likewise, the MIC of L. plantarum K10 in penicillin was higher than that of the reported 46 kinds of isolated L. plantarum strains. The MIC of L. plantarum K10 in gentamicin, streptomycin, clindamycin, erythromycin, vancomycin, and chloramphenicol was similar to the reported 46 kinds of isolated L. plantarum strains. K10 was susceptible to erythromycin and rifampicin according to the European Food Safety Authority (EFSA, 2008) and Scientific Committee for Animal Nutrition (SCAN, 2002). Note, however, that K10 was resistant to gentamycin, kanamycin, streptomycin, ampicillin, ciprofloxacin, tetracycline, vancomycin, and chloramphenicol because MICs were equal to or higher than the breakpoints according to the European Food Safety Authority (EFSA, 2008) and Scientific Committee for Animal Nutrition (SCAN, 2002). These results show that the L. plantarum K10 strain generally has antibiotic tolerance.
The enzyme activities of the L. plantarum K10 strain are shown in Table 3. The strain did not produce β-glucuronidase, a harmful enzyme associated with the inducement of toxins, carcinogenesis, and mutagens (Dabek, 2008); rather, it produced enzymes such as esterase lipase, leucine arylamidase, valine arylamidase, cystimearylamidase, acid phosphatase, naphtol-AS-BI-phosphohydrolase, β-galactosidase, α-glucosidase, β-glucosidase, and N-acetyl-β-glucosaminidase. Notably, the activities of leucine arylamidase and β-galactosidase were 5 degrees. β-galactosidase, α-glucosidase, and β-glucosidase are the useful enzymes. Especially, the strain had β-galactosidase activity that can relieve the symptoms of lactose intolerance because β-galactosidase hydrolyzes lactose to galactose and glucose in milk (de Verse et al., 2003). According to Lee et al. (2015), L. lactis KC24 isolated from kimchi also produced various enzymes, such as acid phosphate, naphtol-AS-BI-phosphohydrolase, and β-galactosidase, but not the toxic enzyme β-glucuronidase. L. plantarum PH04, which has previously been isolated from fecal samples, did not produce β-glucuronidase (Nguyen et al., 2007).
A value ranging from 0 to 2 is assigned to the standard color: zero represents a negative; 5 represents a reaction of maximum intensity. Values 1 through 4 represent intermediate reactions depending on the level of intensity. The approximate activity may be estimated from the color strength: 1 corresponds to the liberation of 5 nanomoles; 2, to 10 nanomoles; 3, to 20 nanomoles; 4, to 30 nanomoles; and 5, to 40 nanomoles or more.
Biogenic amines exist in a wide range of food products such as dairy products, fish products, meat products, wine, beer, vegetables, fruits, nuts, and chocolates (ten Brink et al., 1990). Because of their potential toxicity, the biogenic amine content of various foods has been widely studied (Stratton et al., 1991). Especially, histamine and tyramine may cause different toxicological effects such as migraine, flushing, neurological disorders, headaches, vomiting, nausea, cardiac palpitations, respiratory disorders, hypertension, and blood pressure instabilities (EFSA, 2011; Ladero et al., 2012). It is important to select the strain that does not produce biogenic amines because they are mainly generated by the decarboxylation of the corresponding amino acid through substrate-specific enzymes of the microorganisms present in the food (ten Brink et al., 1990). L. plantarum K10 did not show any biogenic amine formation from any of the precursor amino acids tyrosine, histidine, ornithine, and lysine (Table 4).
| Strain | Biogenic amine production from precursor amino acid | |||
|---|---|---|---|---|
| Tyrosine | Histidine | Ornithine | Lysine | |
| L. plantarum K10 | - | - | - | - |
To be used as probiotic bacteria, the strain should express high tolerance to acid and bile (Lee and Salminen, 1995). Acid and bile tolerance is required for bacterial growth and is involved in the defense mechanisms in the intestine. The bacteria should also survive passage through the stomach as well as survive in food (Lee and Salminen, 1995; Henriksson et al., 1999; Succi et al., 2005).
Fig. 2 shows the growth curves in MRS broth or MRS broth containing 0.3% bile. The Log value of the population after incubation for 7 h without 0.3% oxgall was 9.29, but it was 8.76 with the addition of 0.3% bile. Therefore, the survival rate of L. plantarum K10 in MRS broth containing 0.3% bile was 94.29%. L. plantarum K10 has probiotic potential because a comparatively high percentage of the strain survived in MRS broth containing 0.3% bile salt.
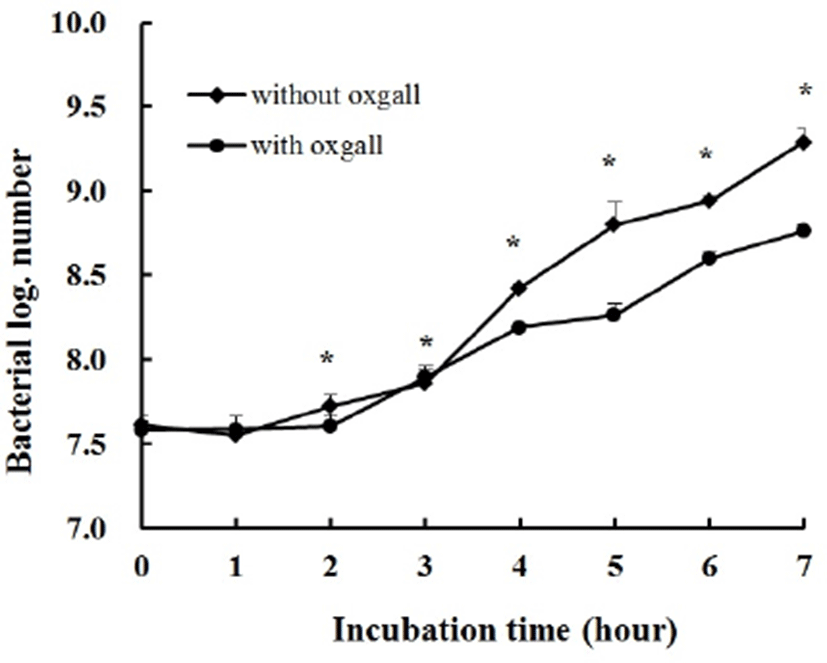
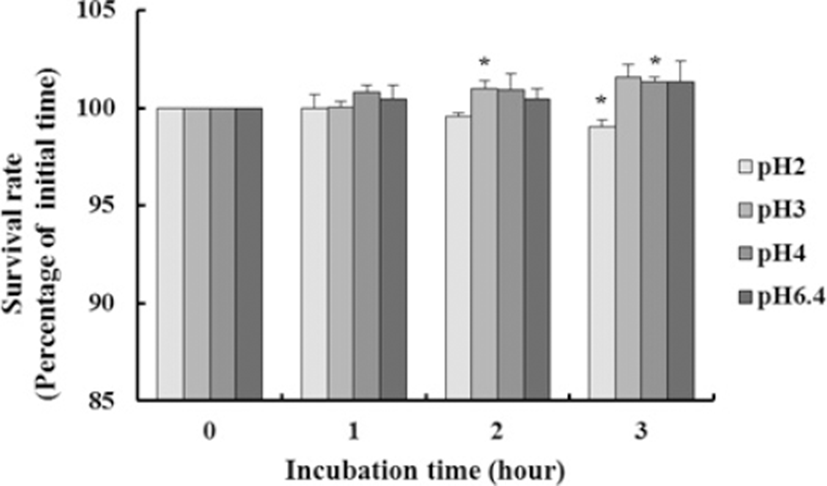
Fig. 3 shows the pH tolerance of L. plantarum K10. It exhibited a 98.93% survival rate after incubation for 3 h in highly acidic conditions (pH 2.0), and the growth of the strain was not influenced by pH 3, 4, and 6.4. In other words, L. plantarum K10 has the best bile and acid tolerance ability because a comparatively high percentage of the strain survived in MRS broth containing 0.3% bile salt as well as under a highly acidic condition.
A similar study was performed by Son et al. (2017). They reported that the viable cell counts of the two strains isolated from kimchi, L. plantarum Ln4 and L. plantarum G72, showed a 1.0 Log CFU decrease after incubation with 0.3% bile salt and did not decrease after incubation at pH 2.5.
Some strains of LAB produce different antimicrobial compounds that can prevent the growth of pathogenic and spoilage bacteria. Antimicrobial metabolites of LAB include hydrogen peroxide, organic acid, bacteriocins, and diacetyl (Ahmadova et al., 2013). Probiotics have to decrease the incidence of pathogenic bacteria for the improvement of health. Therefore, the procedure for selecting probiotics, which are beneficial in the presence of pathogenic bacteria, is important to act against these pathogens (Kesarcodi-Watson et al., 2012).
Table 5 shows the antimicrobial activity of L. plantarum K10 against various pathogenic strains. L. plantarum K10 showed resistance to E. coli, S. Typhimurium, L. monocytogenes, and S. aureus at rates of 90.71, 11.86, 14.19, and 23.08%, respectively. The pH value of pathogens after incubation for 6 h was around 5.98–6.10, but the pH value of a mixed culture with L. plantarum K10 and pathogens was around 4.84–5.25. This means that even lactic acid produced during incubation affected antimicrobial activity, although the effect is not large. Bao et al (2010) investigated the abilities of coaggregation with pathogens of 11 strains isolated from traditional dairy products. The 11 strains showed resistance to E. coli, S. Typhimurium, L. monocytogenes, and S. aureus at rates of 10.5–32.4%, 10.0–29.7%, 11.0–34.0%, and 17.7–49.9%, respectively. The result was similar to L. plantarum K10, but the strain showed high resistance to E. coli.
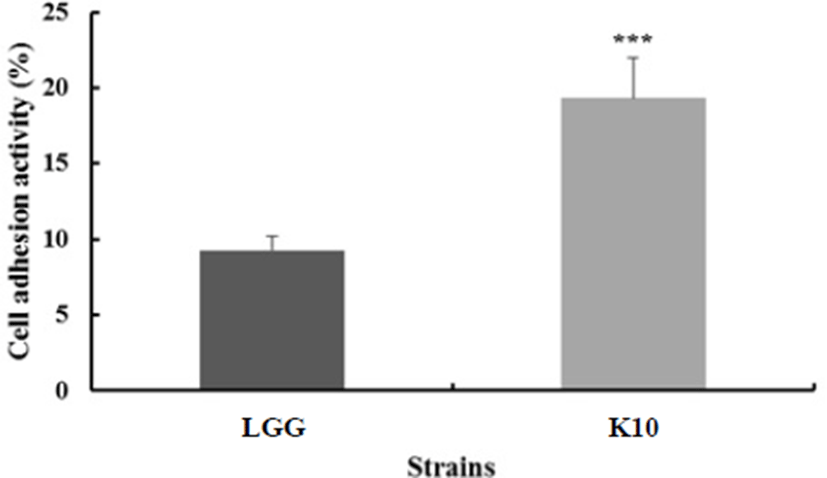
In selecting probiotic strains, their ability to adhere to the intestinal epithelium is one of the main criteria (Blum et al., 1999). This ability is considered a prerequisite to wield beneficial effects, such as exclusion of enteropathogenic bacteria (Bernet et al., 1993) or immunomodulation of the host (Schiffrin et al., 1995). HT-29 cells are derived from colon carcinoma, exhibiting traits of differentiated absorptive enterocytes. Lactobacillus rhamnosus GG was demonstrated to have great ability to adhere to the epithelial cell line in many previous studies (Martín et al., 2005; Gopal et al., 2001). As shown in Fig. 4, 19.34% of L. plantarum K10 adhered to HT-29 cell, and 9.26% of the L. rhamnosus GG strain adhered to the cell. The ability of L. rhamnosus GG to adhere to human intestinal cell line HT-29 was similar to the result of Verdenelli et al. (2009). Thus, we could say that L. plantarum K10 exhibits great adherence to the epithelial surface.
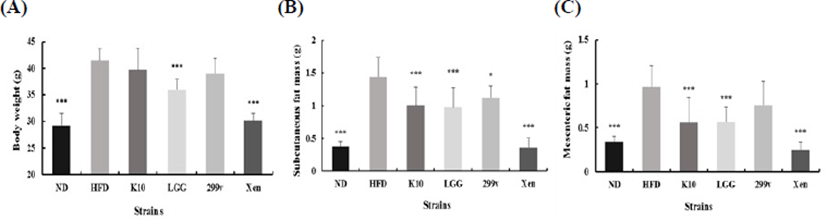
To assess the anti-obesity effect of L. plantarum K10 on the abnormal host status, freeze-dried probiotic strains were incorporated with the IF diet with 5.0×109 CFU/mice/day. L. rhamnosus GG and L. plantarum 299v were selected as reference probiotic strains, and Xenical was chosen for the negative control. After 12 wk, other groups except K10 and 299v showed significantly lower body weight compared to the high-fat diet group (Fig. 5A). Significant weight reduction was not detected on K10 and 299v due to the high variation within the group. Note, however, that the administration of L. plantarum K10 resulted in a reduction of subcutaneous fat mass (Fig. 5B) and mesenteric fat mass (Fig. 5C) compared to the HFD group. In other words, L. plantarum K10 shows an anti-obesity effect on diet-induced obesity mice.
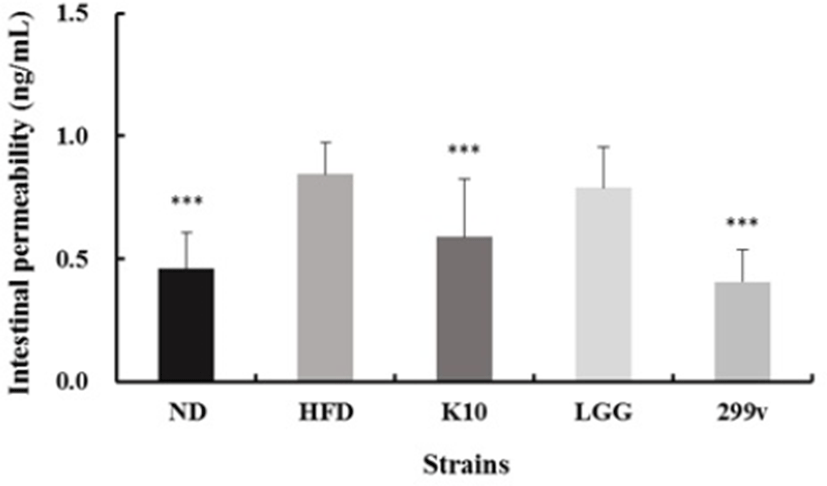
The mucosal cells of the intestine are a single cell layer and are attached to each other by tight junction. Due to changes in the inducement of obesity and alteration in the intestinal microbiota, the cell gap is loosened or damaged, which eventually results in the leaky gut syndrome (Cani et al., 2008). Leaky gut is the result of leaking into the intestinal tract or introduction of polymeric substances directly into the bloodstream of the intestinal tract, resulting in various diseases. If the permeability increases for any reason, pathogen, endotoxin (broken cell wall of gram-negative strains), inflammatory molecules, etc. enter the bloodstream, and intestinal endotoxemia occurs (Thomas et al., 2014). Note, however, that lactic acid bacteria show the effect of reducing intestinal permeability by binding to the intestinal mucosa to form a clustering barrier and regulating the inflammatory conditions to have a positive effect on the intestinal function (Heyman et al., 2005). As shown in Fig. 6, L. plantarum K10 showed improvement in gut permeability compared to the HFD positive control group. Thus, we could say that L. plantarum K10 regulates intestinal permeability.
Conclusion
In this study, we investigated the anti-obesity effect of L. plantarum K10, which was selected from among LAB isolated from kimchi, and studied the physiological characteristics to determine its potential as starter for functional food products.
Based on the nucleotide sequence of 16s rDNA gene, it was named L. plantarum K10. The L. plantarum K10 strain exhibited α-amylase, α-glucosidase, and pancreatic lipase inhibitory activity of 94.66±4.88%, 99.78±0.28% and 87.40±1.41%, respectively, compared with the control. 3T3-L1 cell treated with K10 resulted in about 32.61% reduction in lipid accumulation compared with the untreated control (p<0.05). K10 was resistant to gentamycin, kanamycin, streptomycin, ampicillin, ciprofloxacin, tetracycline, vancomycin, and chloramphenicol. K10 did not show any biogenic amine formation from any of the precursor amino acids. K10 has the best bile and acid tolerance ability. K10 showed resistance to E. coli, S. Typhimurium, L. monocytogenes, and S. aureus at rates of 90.71, 11.86, 14.19, and 23.08%, respectively. K10 exhibited 19.34% adherence to the epithelial surface. As a result of the animal study, K10 showed significantly lower body weight compared to the high-fat diet group. The administration of L. plantarum K10 resulted in a reduction of subcutaneous fat mass and mesenteric fat mass compared to the high-fat diet group. K10 also showed improvement in gut permeability compared to the HFD positive control group. These results suggest its potential as starter for functional food products. In addition, K10 has an anti-obesity effect in 3T3-L1 cell by reducing lipid accumulation. It also reduced body weight in diet-induced obese mice. These results demonstrate that L. plantarum K10 has potential as a probiotic with anti-obesity effects.













