Introduction
Lactic acid bacteria (LAB) have been historically recognized as health promoting bacteria in humans and are widely used as probiotics (de Vos, 2011). In general, LAB exhibit beneficial properties such as anti-diarrheal (Katz, 2006), anticonstipation (Higashikawa et al., 2010), anti-cancer (Riaz Rajoka et al., 2017), and immune stimulation (Tsai et al., 2012), and many LAB are anti-pathogenic bacteria (Sanz et al., 2007). Recently, LAB have attracted interest as bacteria beneficial for gut health since it has been revealed that the gut microbiome is deeply correlated with human physiology (D'Argenio and Salvatore, 2015). Tsai et al. (2014) observed that some LAB strains can modulate the human gut microbiota, which has an influence on the anti-obesity effects as part of metabolic disease prevention. Diets including prebiotics, probiotics, and fermented foods can also modulate the human gut microbiota which has an influence on the enteric and the central nervous systems, resulting in improvement of brain functions (Galland, 2014).
Besides the probiotic effects of LAB, they have been used as starter cultures for various fermented foods including yogurt, sausage, etc. (Ammor and Mayo, 2007; Morelli, 2014) in many different countries throughout the world. Most of the starter cultures used in monoculture fermentations are characterized during fermentation and the processes are well established. However, novel LAB strains for starter cultures that have potential for use in complex food matrices harboring their own indigenous microbial communities are more difficult to follow during the fermentation processes. Reporter proteins such as green fluorescent protein (GFP) and luciferase (Luc) have been used for labeling the bacteria to monitor them in vitro, in situ, and in vivo (Gory et al., 2001; Karimi et al., 2016; Ninomiya et al., 2013; Phumkhachorn et al., 2007). Since these proteins can produce fluorescent and luminescent signals, respectively, in a complex situation, the labeled bacteria can be tracked without culturing for enumeration (Daniel et al., 2015; Yu et al., 2007). Food microbiologists have questioned the behavior of starter cultures or survivability during the fermentation process. In this study, we have tried to track a bioluminescent Lactobacillus plantarum strain during millet seed fermentation where native LAB strains were also cultivated.
Materials and Methods
L. plantarum ULAG11 strain originally isolated from fermented kunu-zaki in Nigeria (Oguntoyinbo and Narbad, 2012) was a gift from Prof. Folarin Anthony Oguntoyinbo working at the Department of Microbiology in University of Lagos (Nigeria). The strain was cultured at 37°C in MRS broth (De Man, Rogosa, and Sharpe; Difco, USA) without shaking. The recombinant L. plantarum ULAG11 (pLuc2) was cultured at 37°C in MRS broth supplemented with 5 μg/mL of erythromycin (Sigma, UK).
Transformation of L. plantarum ULAG11 strain with a recombinant plasmid pLuc2 (Eom et al., 2015), which was developed for lactobacilli bioluminescent labeling, was performed according to the method described in a previously published paper (Moon et al., 2008). The recombinant L. plantarum ULAG11 (pLuc2) strain was selected on the MRS agar plate supplemented with 5 μg/mL of erythromycin (Sigma). Transformation efficiency was 9.6 × 102 cells/μg DNA.
To confirm a luminescent signal from the L. plantarum ULAG11 (pLuc2) strain, a luciferase assay system (E1500; Promega, USA) was used according to the manufacturer’s instructions with modification. Namely, 200 μL of exponential phase culture was centrifuged at 13,000 rpm and resuspended with 20 μL of PBS (phosphate buffered saline; Sigma) and mixed with 100 μL of luciferase assay reagent. The mixture was loaded into a black Costar flat bottom 96-well microplate (Corning, USA) and luminescence was measured by using BMG FLUOstar OPTIMA Microplate Reader (BMG LABTECH GmbH, Germany) according to the manufacturer’s instructions. The parent L. plantarum ULAG11 was used as a negative control.
The L. plantarum ULAG11 (pLuc2) strain was cultured in 100 mL of MRS broth and MRS broth supplemented with 5 μg/mL of erythromycin (Sigma). At the indicated times (0, 2, 4, 6, 8, 10, and 24 h), the cultures were sampled and optical density at 600 nm and luminescence were measured. L. plantarum ULAG11 was cultured in MRS broth alone and used as a negative control.
To investigate luminescence detection sensitivity, 3 different dilution solutions, PBS, MRS broth, and whole milk were tested. L. plantarum ULAG11 (pLuc2), harvested during exponential phase was serially two-fold diluted in each matrix and luminescence was measured as described above.
Millet seeds were soaked in water in a 1:1 ratio (100 g: 100 mL) and set at room temperature overnight. The seeds were washed with water and 100 mL of water was finally added. The mixture was blended and divided into 3 portions. The overnight cultures of L. plantarum ULAG11 and L. plantarum ULAG11 (pLuc2) were separately inoculated in each portion with 1% (v/v) as a final concentration. At the indicated times (0, 3, 6, 9, and 24 h), each 2 mL of the fermented matrix was sampled, and viable cell count, pH, and luminescence were measured.
Results and Discussion
The L. plantarum ULAG11 strain was transformed with pLuc2. The transformation was confirmed by PCR using a specific primer set lucF (5’-TCAAAGAGGCGAACTGTGTG-3’) and lucR (5’-TTTTCCGTCATCGTCTTTCC-3’) targeting a partial region of the luciferase gene in pLuc2. From the PCR, the target PCR product (333 bp) was amplified (data not shown).
Exponential phase L. plantarum ULAG11 (pLuc2) cells cultured in MRS broth produced a luminescent signal measured as 831 RLU (relative luminescence units), whereas the negative control L. plantarum ULAG11 cells did not produce a signal (0 RLU), indicating that the luciferase gene was properly expressed in the recombinant strain (data not shown).
The changes in luminescent intensity during cultivation are shown in Fig. 1. The growth rate of L. plantarum ULAG11 (pLuc2) was slightly lower than that of L. plantarum ULAG11 in MRS broth, but this difference was not significant. The growth rate of L. plantarum ULAG11 (pLuc2) in MRS broth was similar to that in MRS broth supplemented with 5 μg/mL of erythromycin. During culture, the negative control L. plantarum ULAG11 strain produced very low levels of luminescent signals (<10 RLU), whereas the recombinant L. plantarum ULAG11 (pLuc2) produced high levels of luminescent signals (>180 RLU). The highest luminescent signals (2861 and 2858 RLU, respectively) were produced in the mid-exponential phases of L. plantarum ULAG11 (pLuc2) cultured in MRS broth and MRS broth supplemented with erythromycin. Use of antibiotic pressure for improving the luminescent signal did not work in this case. The luciferase structural gene in pLuc2 is linked to the P32 promoter from the pMG36e lactococcal expression vector where the promoter is transcribed constitutively, indicating that enzyme production is dependent on cell growth (Eom et al., 2015; van de Guchte et al., 1989). Cronin et al. (2008) reported that Bifidobacterium breve UCC2003 transformed with pLuxMC3, where the luxABCDE operon is linked to an optimized constitutive promoter (Phelp), produced stable luminescent signals until the stationary phase of growth, with the highest level of the signal in the mid-exponential phase, which is consistent with our result. Additionally, bioluminescent signals of bacterial species harboring the lux operon decline at stationary phase of in vitro culture, which is caused by decreased metabolic activity (Boyandin and Popova, 2003; Cronin et al., 2008; Francis et al., 2001).
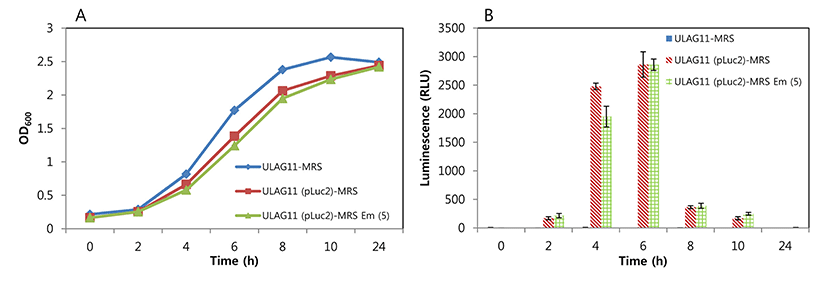
Detection sensitivity of bioluminescent L. plantarum ULAG11 (pLuc2) was the highest in PBS, followed by whole milk and MRS broth, indicating that the matrix is one of the important factors for sensitivity (Fig. 2). The luminescent signals of the two-fold diluted samples in PBS, whole milk, and MRS broth were 111, 78, and 40 RLU, respectively. Even though the slope of trend line for MRS broth was the most gradual, the signal strength was the lowest when compared with those of other samples. These results suggest that for optimal sensitivity of the bioluminescent bacteria from a complex food matrix should be washed and diluted in PBS before the measurement of luminescence.
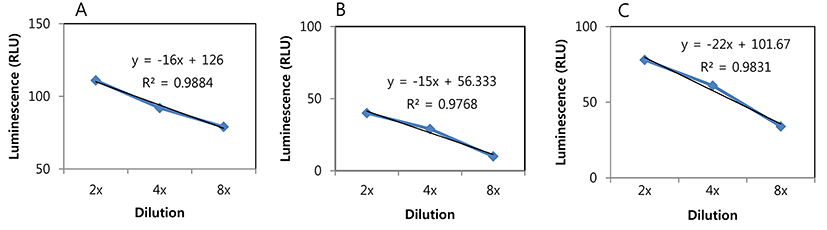
Millet seed is fermented to produce products such as kunu-zaki that is widely consumed by rural African populations. This is a natural fermentation product that contains many different types of endogenous bacteria including LAB (Oguntoyinbo and Narbad, 2012). During millet seed fermentation, only background signals were detected from the millet seed sample without inoculation and with inoculation of L. plantarum ULAG11, whereas the signal from the sample with L. plantarum ULAG11 (pLuc2) was gradually increased until 9 h (Fig. 3A), which is compatible with a gradual increase in the viable cell count of the strain (Fig. 3D). Furthermore, an additional increase in the signal was predicted between 9-24 h since the viable cell count at 24 h was drastically increased (Fig. 3D). As mentioned above, millet seeds had harbored their own LAB strains and the viable cell count during fermentation was increased and it reached a similar level as in the other samples at 24 h (Fig. 3C). Addition of the LAB starter cultures accelerated the drop in pH which is a beneficial fermentation outcome (Fig. 3B).
It is not difficult to track a specific strain in pure culture, but there is difficulty in monitoring the target strain in a complex product derived from mixed fermentation. To differentiate the target strain from the other microorganisms in these fermentations, antibiotic resistance genes can be used for specific detection of recombinant strains based on the culture method. Eom et al. (2008) developed a random integration system using a chloramphenicol acetyltransferase gene for monitoring a Leuconostoc mesenteroides strain as a starter culture in kimchi fermentation, and the strain was successfully monitored during the fermentation period. However, in this case, a pretest for confirming that there is no antibiotic resistant strain in the natural microflora needs to be performed. The culture based method also has some drawbacks since it is laborious and time-consuming. Therefore, other efficient methods for detection of a target bacterial strain, such as a starter culture in complex microflora, are necessary. Bioluminescent labeling of LAB strains can be a solution since most of the natural LAB strains cannot produce a luminescent signal, which enables us to detect only the bioluminescent recombinant target strain. In this study, we applied the labeling system to the L. plantarum strain as a model case and successfully detected it in millet seed fermentation where the natural microflora co-existed. The strategy can be broadly used in food and dairy sciences.













