Introduction
It is established that lactic acid bacteria (LAB) are generally recognized as safe (GRAS) and primarily responsible for the fermentation of food and feed (FAO/WHO, 2002; Sornplang and Piyadeatsoontorn, 2016). As probiotic bacteria, LAB benefit the host by improving the balance of the gut microbial community as well as by providing immunomodulatory effects (Fuller, 1989; Song et al., 2015). In recent years, interest in host health-promoting effects of probiotics has been growing and, as a result, various probiotic products have been developed aimed at improving the health of humans and animals (Heller, 2001).
Lactobacillus is a well characterized genus of the LAB group composed of various species (Claesson et al., 2008). Numerous studies have demonstrated the probiotic potential of various lactobacilli (Ouwehand et al., 2002). Even though there has been a long history of their safe consumption in traditional foods, several criteria should be examined before they are used as probiotic agents or in industrial-grade food products. Probiotic lactobacilli should be resistant to gastric and bile acids, and also colonize the intestinal and/or genital mucosa to persist in the gastrointestinal (GI) tract (Lee et al., 2011). Lactobacilli have been shown to inhibit pathogenic bacteria such as Escherichia coli, Listeria monocytogenes, Salmonella spp., and others. In addition, clinically proven health effects have been reported for lactobacilli, such as cholesterol reduction, diarrhea prevention, reduction of lactose intolerance symptoms, anticancer effects and immunomodulatory effects, all of which are considered functional criteria for probiotic applications (Gareau et al., 2010). Among the lactobacilli, L. acidophilus and L. plantarum are natural and major inhabitants of the GI tract that are often isolated from human and animal biological samples (López-Huertas, 2015). These species have been previously reported to have different probiotic properties (Balasingham et al., 2017); however, their genomes are not yet fully investigated with regard to supporting and elucidating their functionality.
Caenorhabditis elegans as a model system for screening probiotic bacteria has a number of practical advantages; screening is easy to perform, rapid, low cost, and can be scaled up. The body of this nematode is transparent, allowing clear observation of all cells in mature and developing animals (Leung et al., 2008). More importantly, C. elegans has intestinal cells that are similar in structure to human intestinal cells (McGhee, 2007). Recently, our group successfully isolated health-promoting probiotics using C. elegans and we employed a host model in this study to screen potential probiotic bacteria from porcine fecal samples.
The probiotic activity of newly isolated lactobacilli from pig fecal samples was characterized, including acid tolerance, bile tolerance, and adhesion to porcine intestinal epithelial cells. Some of the tested strains exhibited antimicrobial activity and were able to survive for extended periods in a C. elegans host organism. In addition, we determined the complete genome of the L. plantarum strain JDFM LP11, providing genomic evidence for the observed functional activities. The results of this combined analysis demonstrate that the newly identified L. plantarum strain JDFM LP11 hold promise for use as probiotic functional food additives.
Materials and Methods
LAB stains were isolated from fecal samples of healthy piglets (Doozy Farm, Jeollabuk-Do, Korea). Ten grams of fecal sample were diluted in 45 mL L-cysteine solution (8.5 g NaCl, 0.5 g L-cysteine, 1 L distilled water) and plated on de Man, Rogosa, and Sharpe, 0.004% bromocresol purple (MRS-BCP) agar at 37°C for 48 h. Yellow single colonies were selected randomly, sub-cultured in MRS broth, and re-streaked onto MRS agar to ensure purity. Basic assays such as Gram staining and catalase tests were performed according to Bergey's Manual of Determinative Bacteriology (Buchanan and Gibbons, 1974). All isolated LAB were maintained at –80°C in MRS broth containing glycerol solution for further studies. All strains were sub-cultured three times prior to experimental analysis.
The C. elegans strain used in this study was CF512 fer-15(b26)II;fem-1(hc17)IV (Kim and Mylonakis, 2012). It was routinely maintained on nematode growth medium (NGM) plates seeded with E. coli OP50 as described previously (Brenner, 1974). In order to evaluate whether the probiotic candidates had an effect on host lifespan, nematode lifespan was measured according to previously described methods (Aballay et al., 2000; Garigan et al., 2002) with modifications. Groups of twenty worms were exposed to each probiotic candidate on NGM plates and the numbers of live worms were scored every day. To correctly determine scores, worms were transferred to fresh plates every three days. A worm was regarded as dead when it failed to respond to a gentle touch. All C. elegans were cultured at 25°C. In addition, to screen for colonization of the C. elegans intestinal tract, the numbers of bacterial cells in worm intestines were measured according to previously described methods with slight modifications (Garsin et al., 2001). Following exposure of C. elegans to individual probiotic candidates on NGM plates containing nystatin (5 μg/mL) for five days, ten worms were picked at random, washed twice in M9 buffer, and placed on brain heart infusion (BHI) plates containing both kanamycin (100 μg/mL) and streptomycin (100 μg/mL). These plates were exposed to 5 μL gentamicin solution (25 μg/mL) for 5 min, and results were compared to L. rhamnosus GG as a positive control. Subsequently, worms were washed five times with M9 buffer, transferred into a 1.5 mL Eppendorf tube containing M9 buffer with 1% Triton X-100, and mechanically disrupted using a pestle (Kontes, Vineland, NJ). The resulting diluted worm lysates were plated on modified MRS (pH 5.0) agar and incubated at 37°C for 48 h.
Tolerance to low pH and bile content was assessed as described by Jacobsen et al. (1999), with minor modifications. The ability of the strains to grow at low pH was evaluated in acidified MRS broth (final pH 2.5) containing 1,000 unit/mL of pepsin (Sigma, St. Louis, MO, USA). The tolerance of the strains to bile was determined in MRS broth containing 0.5% oxgall (Sigma, USA). Ten milliliters of each modified MRS was inoculated with a bacterial suspension to a final cell concentration of approximately 1.0×107 CFU/mL. Acid tolerance was evaluated by measuring survival after 3 h-incubation at 37°C. Bile tolerance was evaluated by measuring survival after 24 h-incubation at 37°C.
The ability of the strains to adhere to porcine epithelial cells was investigated according to the method of Kim et al. (2009) with slight modification. Monolayers of intestinal porcine epithelial cell lines IPEC-J2, derived from pig small intestine were prepared in RPMI-1640 medium (Sigma-Aldrich, St. Louis, Mo, USA) supplemented with 10% fetal bovine serum (FBS) (Sigma-Aldrich, St. Louis, Mo, USA) in 24-well tissue culture plates (BD Biosciences, San Jose, CA, USA) at a concentration of 4×104 cells/well. The cells were incubated with approximately 1×107 CFU/mL of the strain to be tested. After 2 h-incubation at 37°C, the monolayers were washed six times with PBS. Adherent bacteria were detached by repeatedly pipetting with chilled sterile water, diluted in PBS solution and then counted on MRS agar plates.
The inhibitory effects of the lactobacilli on intestinal pathogens was measured using an overlay method. Two representative porcine intestinal pathogens including pathogenic E. coli and Salmonella Typhimurium were used for the test. Bacteria were grown in Luria-Bertani (LB) broth (Difco, USA) at 37°C for 18 h. Briefly, 10 μL aliquots of overnight liquid cultures of LAB isolates were dropped onto fresh MRS agar plates. Seeded plates were incubated at 37°C for 48 h and subsequently overlaid by soft medium, containing 0.8% agar and inoculated with overnight liquid cultures of pathogens which had been incubated for 18–24 h at 37°C. Inhibitory activity was observed as a zone of clearance around the colony of the LAB isolate.
Chromosomal DNA from each strain was extracted and the 16S rRNA gene was amplified using universal primers. The PCR primer sequences were as follows: forward primer, 5´-AGAGTTTGATCCTGGCTCAG-3´; reverse primer, 5´-GGTTACCTTTGTTACGACTT-3´ (Bioneer, Korea). The thermal cycling parameters were denaturation at 94°C for 5 min, followed by 30 cycles of denaturation at 94°C for 1 min, annealing at 50°C–55°C for 1 min, polymerization at 72°C for 40 s, and a final polymerization step at 72°C for further 5 min. Amplified products were purified for sequencing using a Gel Extraction Kit (Intron, Korea). The sequences of the final products were analyzed using an ABI 377 automated DNA sequencer (Perkin Elmer, USA). Sequence homologies were examined by comparing the obtained sequences with those in DNA databases (http://www.ncbi.nlm.nih.gov/BLAST).
Genomic DNA of the strain JDFM LP11 was obtained using an UltraClean Microbial DNA Isolation Kit (MoBio, Carlsbad, CA, USA) as described in the manufacturer’s protocol. The extracted DNA concentration and quality were measured using an Optizen™ NanoQ spectrophotometer (Daejeon, Korea). An 8–12 kb library was prepared following the Pacific Biosciences manual; sequencing procedure and filtration/de-novo assembly of raw sequence data was carried out on the PacBio RS II system (Pacific Biosciences, Menlo Park, CA, USA) using C4 chemistry on single molecule, real-time cells with a 120 min sequence capture protocol according to the PacBio Hierarchical Genome Assembly Process using the Quiver software package. Assembled contigs, including short lengths (< 20,000 bps) and low coverage (< 50×) were sorted for further experiments. The annotation of the JDFM216 genome was performed using clusters of orthologous groups (COG) annotation and evolutionary enealogy of genes: non-supervised orthologous groups (EggNOG) categorization was conducted using previously described methods (Jang et al., 2017; Wu et al., 2011). For comparative genome analysis, complete genome sequences of L. plantarum were searched on the Ensemble bacteria database (http://bacteria.ensembl. org/index.html). Average nucleotide identity (ANI) values were calculated for all eight listed strains (Richter and Rosselló-Móra, 2009). OrthoFinder (v1.1.10) and PRANK (Löytynoja and Goldman, 2008) methods were used to determine ortholog genes for the eight complete genomes and for multiple sequence alignment of each ortholog gene, respectively. In addition, the construction of phylogenetic trees by the neighbor joining method were performed using the R program and JSpecies web server. These sequences were converted into corresponding cDNA sequences using PAL2NAL (Suyama et al., 2006) and poorly aligned transcripts were eliminated using Gblocks (Castresana, 2000). After all filtering steps were performed, a total of 2,449 orthologs remained. Moreover, PAML4 analysis by the maximum likelihood method (Yang, 2007) was used to estimate dS (the synonymous substitution rate) and dN (the nonsynonymous substitution rate), as well as phylogenetically featured genes investigated by the branch and branch-site models.
Nematode survival was analyzed using the Kaplan-Meier method and differences were determined using the log-rank test (STATA6; STATA, College Station, TX, USA). Student's t-test was performed to determine statistical differences in CFU counts for determining bacterial counts. All data represent the results of three independent measurements. A p value of 0.05 in all replicate experiments was considered a significant difference compared to the control.
Results and Discussion
Approximately 250 LAB strains isolated from piglet feces were investigated by assessing in vivo colonization of the intestinal tract and lifespan extension using C. elegans as a model. Among them, JDFM LP11 was finally selected as having the best probiotic characteristics. As shown in Fig. 1, JDFM LP11 showed remarkably high colonization ability of the intestinal tract of the C. elegans host. This strain exhibited an outstanding persistence on the C. elegans intestinal wall at over 4.3 CFU/mL/worm; this is higher than the value of 3.2 CFU/mL/worm obtained for the positive control, L. rhamnosus GG, which has been shown to bind to enterocytes in several previous studies (Fig. 1A).
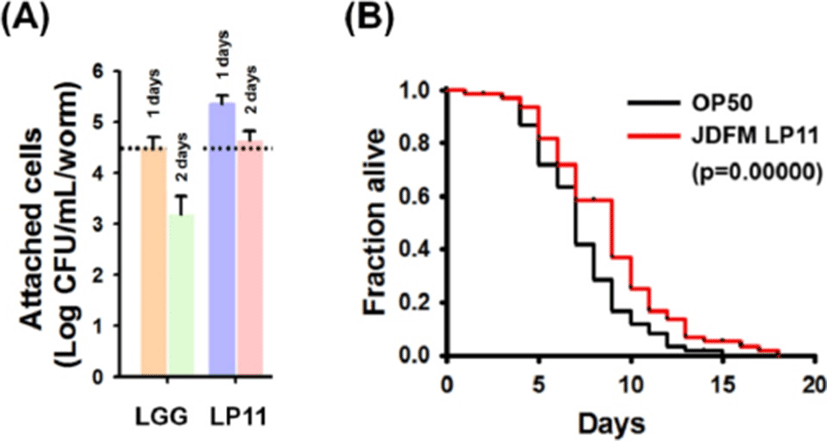
Equally importantly, worms exposed to the JDFM LP11 had a significantly extended lifespan, as shown in Fig. 1B. In particular, JDFM LP11 notably prolonged the average lifespan compared to worms fed with E. coli OP50 (normal feed for C. elegans). Hence, JDFM LP11 demonstrated host health-promoting effects as shown by its anti-aging activity, as well as showing excellent colonization of the worm intestinal tract.
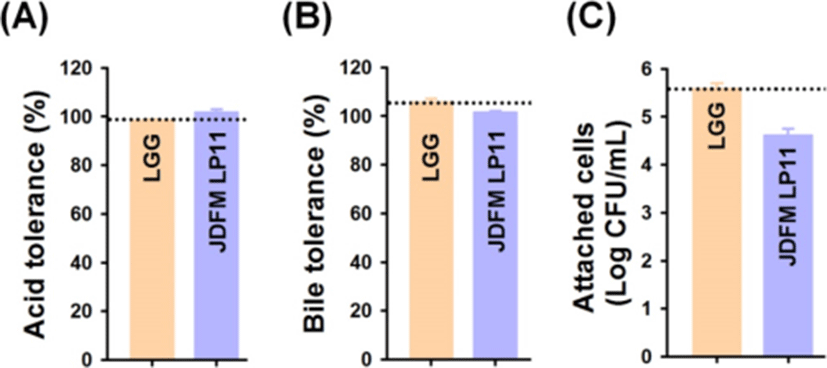
We assessed the isolated JDFM LP11 using conventional in vitro methods. Tolerance to low pH and bile salts, which reflect conditions in the digestive tract, are fundamental to probiotic activity. The ability of the JDFM LP11 to survive in acidic conditions (pH 2.5), enabling them to survive in the large intestine of humans, was examined by incubating isolates for 3 h at 37°C with 1,000 unit/mL pepsin and viable cell counting. Even though exposure of acidic conditions, JDFM LP11 achieved a high survival rate similar with L. rhamnosus GG as control. And then, this strain was also examined for its ability to grow at concentrations of 0.5% bile salts for 24 h. Survival rates of the positive control and JDFM LP11 strain was approximately 100%. This isolate therefore exhibits good viability and resistance to acidic conditions and bile salts (Fig. 2A and B). In addition, we examined the ability of the selected strains to persist in the gut environment and achieve in vivo colonization by using IPEC-J2 cells. JDFM LP11 adhered strongly to the IPEC-J2 cells (≥ 4.5 log CFU/mL), although exhibiting lower adherence than the positive control L. rhamnosus GG (Fig. 2C). The cell adhesion number of JDFM LP11 was consistent with the attachment ability observed using C. elegans in vivo (Fig. 1A). Previously, we reported that C. elegans nematode could provide on the simple and reliable intestinal environment and attachment results of probiotic strains including L. rhamnosus GG (that possesses profound attachment ability both in vivo and in vitro assays) using C. elegans intestinal tract were highly correlated with that of in vitro assays including mucus layer and intestinal cell lines (Park et al., 2014). Taken together, JDFM LP11 selected by our direct, in vivo methods also showed characteristic probiotic properties with in vitro biological assays using an intestinal cell line.
In addition, inhibition of pathogenic bacterial strains including E. coli and Salmonella spp. by isolated JDFM LP11 was also observed on this study (data not shown). Based on 16s rRNA sequencing, JDFM LP11 was identified as L. plantarum and was subjected to further genome analysis.
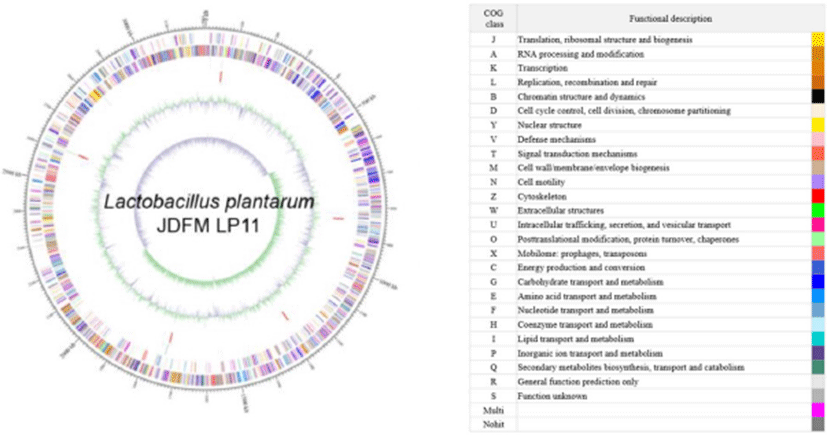
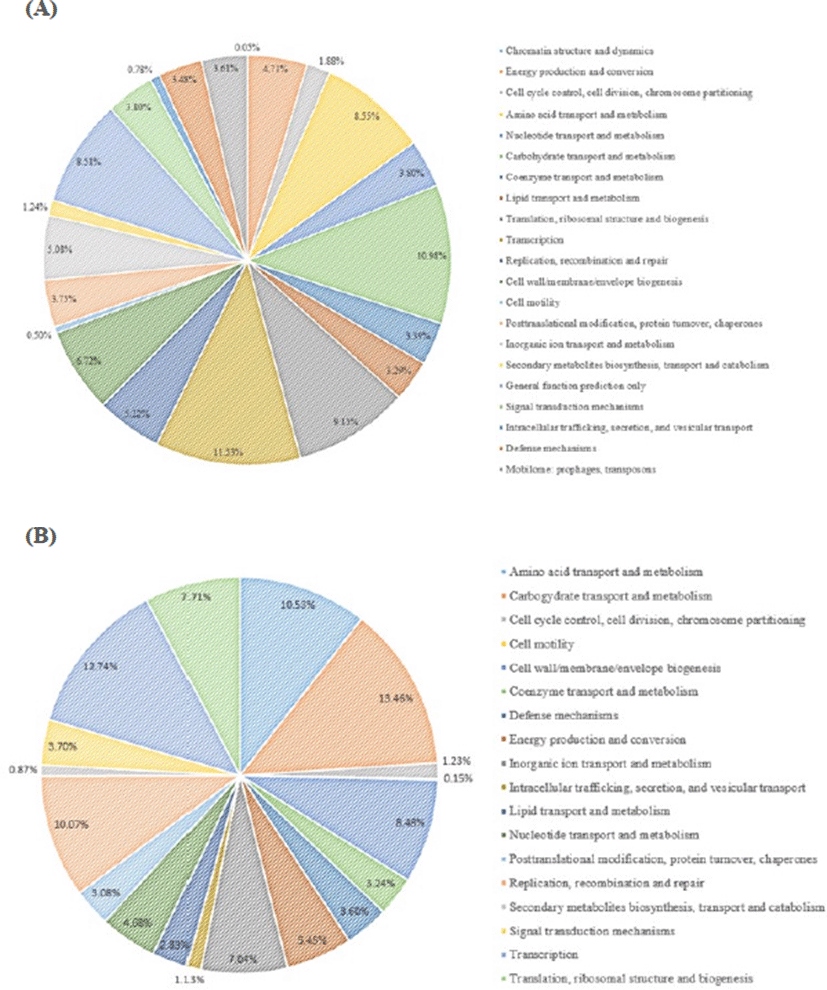
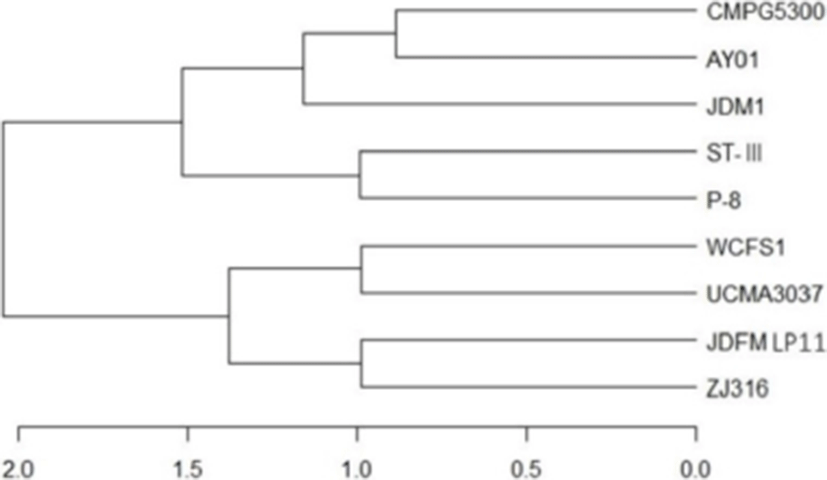
The L. plantarum JDFM LP11 genome possesses a single, circular DNA chromosome of 3,206,883 bp. The JDFM LP11 genome has a total of 3,021 open reading frames (ORFs) as well as 82 non-coding genes (Fig. 3 and Table 1). Importantly, the ORFs of JDFM LP11 were smaller in number than the L. fermentum strains, even though genome size was similar. Based on our bioinformatics analysis, no plasmid could be detected in JDFM LP11, similar to other strains. As shown in Fig. 4, the predicted ORFs were grouped by COG functional and EggNOG subsystem categorizations. First, COG functional categorization showed that 1,472 ORFs (48.72% of the COG assigned ORFs) belonged to five major COG functional categories including amino acid transport and metabolism, carbohydrate transport and metabolism, translation, ribosomal structure and biogenesis, recombination, replication, repair, and general function prediction (Fig. 4A). In addition, EggNOG categorization mainly classified ORFs into those responsible for amino acid transport and metabolism, carbohydrate transport and metabolism, cell wall/membrane/envelope biogenesis, and replication/recombination/repair as well as transcription (55.28% in a whole population of 3,021 ORFs; Fig. 4B). Next, we conducted an ANI, comparative tree analysis of the strain JDFM LP11 compared with genome sequences of the same species. The ANI trees were constructed from the eight available complete genome sequences in the Ensemble bacteria database (Fig. 5). Three strains, namely WCFS1, UCMA3037, and ZJ316, grouped together with JDFM LP11. Subsequently, dN/dS analysis in a branch-site model was performed to identify the phylogenetically featured genes in the JDFM LP11 strain. Three phylogenetically featured genes and their amino acid sequence changes were identified in this study (Table 1 and 2); putative endopeptidase p60, lichenan permease IIC component, and protein TsgA, which are an endopeptidase, a component of the phosphotransferase system (PTS), and a major facilitator in the gut environment, respectively. It has been established that proteolytic enzymes are inhibited in vivo by protein inhibitors produced by symbiotic bacteria in our gut to protect themselves from digestive peptidases (Garcia-Ferrer et al., 2015). Recently, endopeptidase LytA has been shown to play a key determinant role in the morphology of L. plantarum as well as having featured roles in both cell separation and autolysis under stress conditions (Rolain et al., 2012); hence we consider that putative endopeptidase p60 may be responsible for survival of L. plantarum JDFM LP11 in the gut environment and interactions with gut microbiota. In addition, carbohydrate transport and metabolism in bacteria may depend on a functional PTS (Postma et al., 1993). In the present study, we found phylogenetically featured genes encoding a lichenan permease IIC component. Lichenan (or lichenin) is a linear polysaccharide isolated from lichen; thus lichenase (formerly 1,3-1,4-β-glucanase) and its permeability system is important for utilization of lichenan (Chaari et al., 2012). Importantly, supplementation of this enzyme produced by Clostridium and Enterococcus can increase β-glucan digestibility in feed components, improve feed conversion efficiency, and reduce sanitary problems in the animal feed industry. Furthermore, lichenase and lichenan-related PTS are employed as targets for selective growth of animal beneficial intestinal microbiota including bifidobacteria and lactobacilli as non-nutritional food additives. Given this information, it is reasonable to assume that L. plantarum JDFM LP11 utilizes various prebiotic substrates through these PTS components. In addition, homocysteine S-methyltransferase, as a key player in methionine metabolism (Obeid, 2013), was identified in the present study. In addition, TsgA is one of major facilitator superfamily (MFS) and which are induced by tellurite and selenite in E. coli (Pao et al., 1998). Interestingly, MFS involved in TsgA is present ubiquitously in bacteria and archaea. Unfortunately, there is no information for TsgA in the literature as a transporter in the genus Lactobacillus. In conclusion, we suggest that the featured genes may positively influence the colonization and survival of L. plantarum JDFM LP11 and its interaction with commensal microbiota or pathogens in the gut.