Introduction
Several recent studies have demonstrated that non-aureus staphylococci (NAS), including coagulase-negative staphylococci (CoNS), have a substantial impact on human and animal health (Fisher et al., 2018). Nosocomial CoNS are particularly well-known for their ability to form biofilms and acquire resistance to multiple antimicrobial agents (Nadell et al., 2009; O’Gara and Humphreys, 2001; Schilcher and Horswill, 2020). It has been suggested that NAS in livestock farm environments represent a significant reservoir of antimicrobial resistance (AMR) genes, facilitating the horizontal transfer of AMR (Feßler et al., 2018; Shen et al., 2013). Methicillin-resistant Staphylococcus aureus (MRSA) has also been proposed to have acquired staphylococcal cassette chromosome mec (SCCmec) from CoNS, which normally colonize various animal hosts.
The high prevalence of antimicrobial-resistant staphylococci, particularly multidrug-resistant (MDR) staphylococci, in livestock animals and farm environments has become a significant hazard to food safety and public health. Although a relatively large number of studies have focused on livestock-associated MRSA (LA-MRSA) in swine and cattle farms, the prevalence of NAS and their AMR profiles in livestock farms, especially broiler farms, are not well established. Only a few recent studies have investigated the prevalence of antimicrobial-resistant S. aureus in broiler farms, poultry slaughterhouses, and retail chicken meat samples in Korea (Kim et al., 2018; Lee et al., 2022). In addition, NAS species, such as Staphylococcus agnetis, Staphylococcus saprophyticus, Staphylococcus xylosus, Staphylococcus simulans, and Staphylococcus lentus, have been reported as opportunistic pathogens in poultry (Boamah et al., 2017; Pyzik et al., 2019; Silva et al., 2022; Szafraniec et al., 2020). However, information on the distribution of NAS species in broiler farms and their AMR profiles is still not available in Korea.
In the present study, we analyzed the species profiles of NAS isolates collected from broiler farms, including healthy broilers, farm environments, and farm workers. In addition, the AMR profiles of the NAS isolates and the major genetic factors involved in the resistance phenotypes, especially methicillin and fluoroquinolone resistance, were examined. To the best of our knowledge, this is the first study to report the species profiles, AMR, and genetic factors associated with fluoroquinolone resistance in NAS isolates from broiler farms in Korea.
Materials and Methods
All swab samples were obtained from 20 different broiler farms across eight provinces of Korea in 2019 as previously described (Lee et al., 2022). A total of 916 swab samples were collected from healthy broilers (n=782; cloacal and throat swabs), farm facilities (n=71; floor, fence, drinking system, feeder pan, sewage, and ventilators), and farm workers (n=63; hand and nasal swabs). After sampling, all swabs were transported using ice-cooled containers and processed for the isolation of NAS within 36 h of sample collection. The sampling protocol was reviewed and approved by the IRB/IACUC at Chung-Ang University.
Isolation and culture of NAS strains from swab samples were performed as previously described, with minor modifications (Lee et al., 2022). Briefly, all swab samples were inoculated into 4.5 mL of tryptic soy broth (Difco Laboratories, Detroit, MI, USA) containing 10% sodium chloride and incubated at 37°C for 17–24 h for enrichment. Next, 15–20 μL of the pre-enriched cultures were streaked onto Baird-Parker agar (Difco Laboratories) supplemented with egg yolk and potassium tellurite (Becton Dickinson, Sparks, MD, USA) and incubated for 48 h at 37°C. One or two colonies of presumptive staphylococci per sample were then selected for subsequent identification of species. The species of the NAS strains were first identified by using a matrix-assisted laser desorption ionization (MALDI)-Biotyper classification system (Bruker, Bremen, Germany) and then 16S rRNA sequencing for strains with <2.0 scores in the MALDI-Biotyper analysis, as previously described (Yang et al., 2022).
To determine the AMR profiles of the NAS strains, standard disc diffusion assays were performed according to the Clinical and Laboratory Standards Institute guidelines (CLSI) (CLSI, 2020a; CLSI, 2020b) and the European Committee on Antimicrobial Susceptibility Testing (EUCAST). The 14 antimicrobial agents included in the disc diffusion assays were ampicillin (AMP, 10 μg), cefoxitin (FOX, 30 μg), penicillin (PEN, 10 units), gentamicin (GEN, 50 μg), chloramphenicol (CHL, 30 μg), ciprofloxacin (CIP, 5 μg), clindamycin (CLI, 2 μg), erythromycin (ERY, 15 μg), fusidic acid (FUS, 50 μg), mupirocin (MUP, 200 μg), rifampin (RIF, 5 μg), sulfamethoxazole-trimethoprim (SXT, 23.73–1.25 μg), quinupristin-dalfopristin (SYN, 15 μg), and tetracycline (TET, 30 μg). All antimicrobial discs were purchased from BD BBLTM (Becton Dickinson, Franklin Lakes, NJ, USA), except for the MUP discs (Oxoid, Hampshire, UK). The minimum inhibitory concentrations of vancomycin (VAN), tigecycline (TGC), linezolid (LZD), and teicoplanin (TEC) were determined using standard E-test strips (bioMérieux, Craponne, France) on Mueller-Hinton agar (Difco Laboratories). Two reference strains, S. aureus ATCC29213 and S. aureus MW2, were used in antimicrobial susceptibility assays.
All NAS strains displaying resistance phenotypes to β-lactam antimicrobial agents were subjected to polymerase chain reaction (PCR) to detect mecA gene, as previously described (Geha et al., 1994). The types of SCCmec were then determined in all mecA-positive NAS strains using previously described multiplex PCR methods that amplify chromosomal cassette recombinase (ccr) genes and mec regulatory elements. The combinations of ccr types and mec complexes were analyzed to assign the SCCmec types of methicillin-resistant NAS strains (Geha et al., 1994; Mendoza et al., 1998).
To detect the mutations in fluoroquinolone-resistant strains, quinolone-resistance determining regions (QRDRs) within gyrA, gyrB, parC, and parE were PCR-amplified using specific primer sets as previously described (Lee et al., 2020; Sreedharan et al., 1991; Takahashi et al., 1998; Takahata et al., 1997). The PCR products of the QRDRs were then sequenced (Bionics, Seoul, Korea), and mutations were identified using the Cluster Omega server (http://ebi.ac.uk/Tools/msa/clustalo/). The published sequences of S. agnetis (CP009623.1), Staphylococcus chromogenes (CP046028.1), Staphylococcus arlettae (AP019698.1), Staphylococcus cohnii (CP027422.1), Staphylococcus condimenti (CP018776.1), S. haemolyticus (CP035291.1), Staphylococcus gallinarum (CP086207.1), S. saprophyticus (CP031196.1), S. simulans (LT963435.1), and S. xylosus (CP013922.1) were used to design QRDR-specific primer sets in each species (Supplementary Table S1).
Results
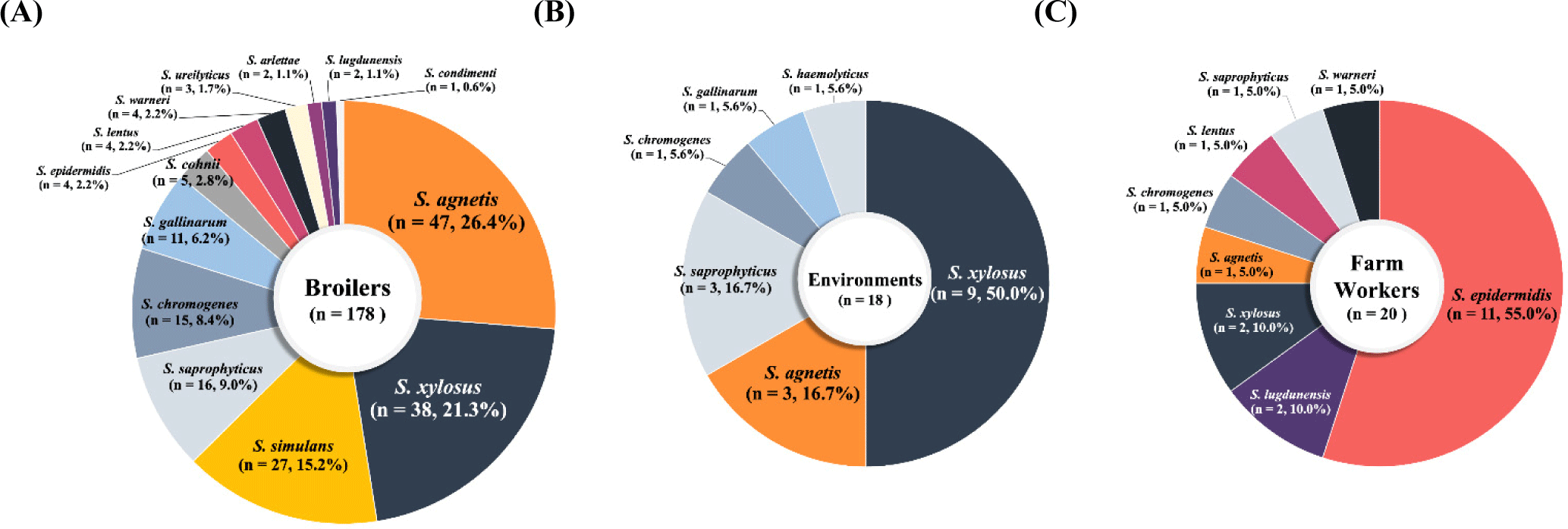
A total of 216 NAS strains of 16 different species were isolated from 916 swab samples (216/916, 23.58%) collected from broiler farms during the study period (Figs. 1A, B, and C). Of the 216 NAS strains, 148 (68.5%) CoNS strains displayed 14 different species diversities, whereas 68 coagulase-variable staphylococci (CoVS) were composed of only two species of staphylococci, S. agnetis and S. chromogenes. As shown in Fig. 1A, 178/216 (82.4%) NAS strains were isolated from healthy broilers, with S. agnetis and S. xylosus being most frequently associated with healthy broilers (Fig. 1A). Similar to the profiles of NAS strains in broilers, 83.3% (15/18) of NAS strains from farm environments were S. xylosus (n=9), S. agnetis (n=3), and S. saprophyticus (n=3) strains (Fig. 1B). NAS strains from farm workers were distinctive from those of broilers and farm environments in that Staphylococcus epidermidis (11/20, 55%) was most prevalent in the nasal cavities of farm workers (Fig. 1C).
Six methicillin-resistant NAS were identified in 216 NAS strains (3.24%) isolated from broiler farms (Table 1). All six strains were mecA-positive (Table 1) and were resistant to FOX (Table 2). Analysis of SCCmec elements in seven methicillin- resistant NAS revealed that two CoNS (S. epidermidis and S. haemolyticus) carried SCCmec V (Table 1). Two methicillin-resistant S. epidermidis strains isolated from farm workers contained SCCmec II and SCCmec IV. Although two CoNS strains (Staphylococcus sciuri and Staphylococcus ureilyticus) were mecA-positive, the SCCmec types could not be determined using PCR because of unidentifiable ccr or mec types.
NAS, non-aureus staphylococci; AMP, ampicillin; FOX, cefoxitin; PEN, penicillin; CHL, chloramphenicol; CIP, ciprofloxacin; CLI, clindamycin; ERY, erythromycin; FUS, fusidic acid; GEN, gentamicin; MUP, mupirocin; RIF, rifampin; SXT, trimethoprim-sulfamethoxazole; SYN, quinupristin- dalfopristin; TET, tetracycline; VAN, vancomycin; TEC, teicoplanin; LZD, linezolid; TGC, tigecycline; MDR, multi-drug resistance; CoVS, coagulase-variable staphylococci; CoNS, coagulase-negative staphylococci.
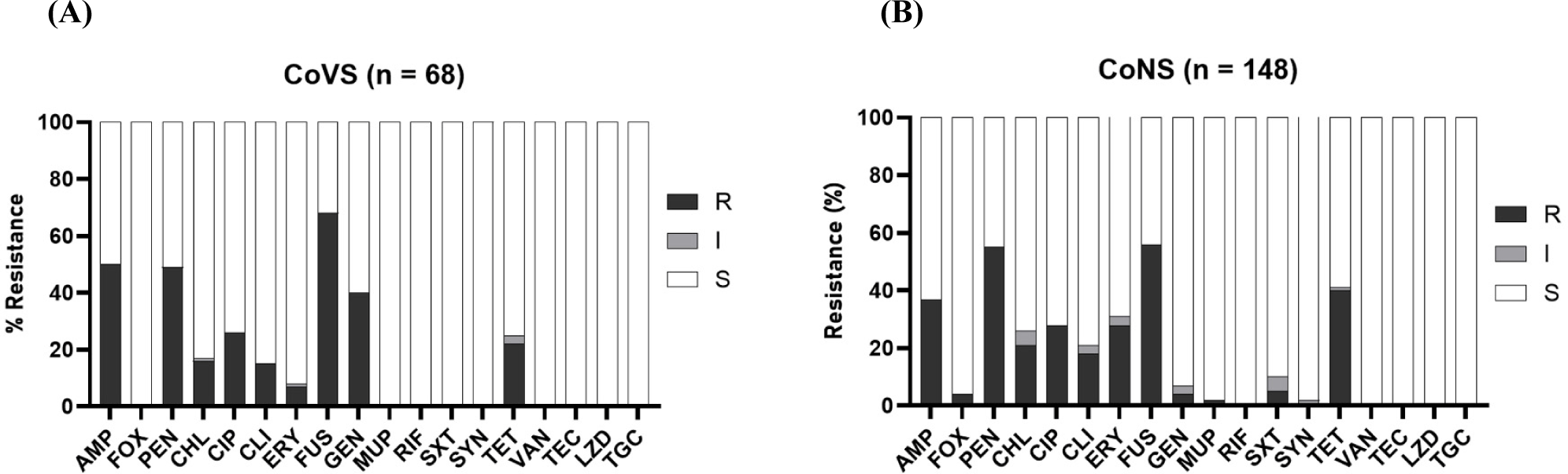
All 216 NAS isolates (both CoVS and CoNS) were susceptible to RIF, VAN, LZD, TEC, and TGC (Figs. 2A and B). CoVS strains (41.2%), particularly S. agnetis strains (54.9%), showed higher levels of resistance to GEN than the CoNS strains (4.1%; Table 2). Both CoVS and CoNS displayed relatively higher levels of resistance to FUS, AMP, PEN, TET, and CIP than to other antimicrobial agents. MDR phenotypes, which show resistance to more than three different classes of antimicrobial agents, were observed in 55.9% and 44.6% of CoVS and CoNS strains, respectively (Table 2). When the two major species of CoVS (S. agnetis and S. chromogenes) were compared for resistance, S. agnetis strains exhibited significantly higher levels of resistance to AMP, PEN, FUS, and GEN, whereas S. chromogenes showed higher resistance to CIP, CLI, and TET (Table 2). Among the four major groups of CoNS strains, S. xylosus and S. saprophyticus strains displayed higher levels of MDR than S. simulans and S. epidermidis strains. S. xylosus and S. saprophyticus strains were highly resistant to PEN (>60%), FUS (>79.6%) and TET (>42.9%).
Eighteen CoVS (18/68 strains, 26.5%) and 39 CoNS (39/148 strains, 26.4%) strains displayed resistance to fluoroquinolones (Table 3). All 57 fluoroquinolone-resistant NAS strains had at least one point mutation in the QRDRs of gyrA, gyrB, parC, or parE genes. As shown in Table 3, all fluoroquinolone-resistant NAS strains had the S84L mutation in gyrA, except for six strains of S. cohnii (n=1), S. condimenti (n=1), S. lentus (n=3) and S. epidermidis (n=1). Mutations in gyrB were identified in only three species of CoNS: S. sciuri, S. simulans, and S. xylosus strains. Among the four point mutations in gyrB (S567A, A512R, E490G, and S494T), the S567A mutation was most frequently identified in fluoroquinolone-resistant CoNS, especially in S. xylosus. Although the S80L mutation was most frequently observed in fluoroquinolone-resistant NAS strains, particularly in S. agnetis and S. saprophyticus strains, high levels of heterogeneity were observed in parC mutations of S. epidermidis, S. gallinarum, and S. xylosus strains (Table 3). In contrast to mutations in gyrA, gyrB, and parC, which were identified in multiple species of fluoroquinolone-resistant NAS, the T379K mutation in parE was detected only in S. simulans strains.
Discussion
Staphylococci are normal flora on the skin and mucous membranes of poultry and other livestock animals. However, some staphylococci can cause opportunistic infections in poultry and food poisoning in humans (Lee et al., 2022; Mekhloufi et al., 2021; Silva et al., 2022). Although recent studies have demonstrated the important role of CoNS and non-aureus CoPS in the transmission of AMR between livestock animals and humans (Huebner and Goldmann, 1999; Silva et al., 2022), limited data are available on the prevalence and species profiles of NAS, as well as their AMR resistance profiles.
Overall, the prevalence rates of CoVS and CoNS among the 216 NAS strains collected from broiler farms were 31.5% (68/216) and 68.5% (148/216), respectively (Table 1). Previous studies conducted in Korea (Lee et al., 2020) and other countries (Marek et al., 2016; Osman et al., 2016; Pyzik et al., 2019) also reported higher prevalence of CoNS compared with CoVS in broiler and retail chicken meat. In a study by Awan and Matsumoto (1998), 77/79 staphylococcal isolates from broilers that showed signs of illness were CoNS, such as S. simulans, S. lentus, and S. cohnii. These results suggest that NAS strains, especially various species of CoNS strains, are colonizing healthy broilers and farm environments, causing opportunistic infections in broilers. In this study, although only two species of CoVS, S. agnetis and S. chromogenes, were detected, 14 different species of CoNS were identified (Table 1 and Figs. 1A, B, and C). The most frequently isolated species of NAS from healthy broilers were S. agnetis (26.4%), S. xylosus (21.3%), and S. simulans (15.2%; Fig. 1A). Frequent occurrence of S. agnetis (3/18, 16.7%) and S. xylosus (9/18, 50.0%) were also observed in the farm environment (Fig. 1B). Recently published studies by our group and others reported that various NAS strains present in retail chicken meat (Martins et al., 2013; Osman et al., 2016; Yurdakul et al., 2013). In particular, the overall detection rate of NAS in retail chicken was 45.3% in Korea, and the most frequently detected NAS species in retail chicken meat samples were S. agnetis (19.4%) and S. saprophyticus (19%; Lee et al., 2020), suggesting S. agnetis may have transmitted from broiler farms to retail chicken meat. Although the most common NAS species in the nasal cavities of farm workers in this study were S. epidermidis (11/20, 55.0%) and Staphylococcus lugdunensis (2/20, 10.0%), a few strains of NAS species identified in broilers, such as S. agnetis (n=1), S. xylosus (n=2), and S. saprophyticus (n=1), were also isolated from the broiler farm workers (Fig. 1C). These NAS species have been shown to be mainly associated with local and chronic infections in broiler chickens and other poultry (Marek et al., 2016; Szafraniec et al., 2020). It has been reported that S. agnetis and S. xylosus have various virulence factors involved in host cell adherence, immune evasion, and toxin biosynthesis, which contribute to the persistence and pathogenesis of staphylococci in poultry (Szafraniec et al., 2020). S. agnetis has been associated with bacterial chondronecrosis, endocarditis, and septicemia in broiler chickens (Szafraniec et al., 2020). Moreover, exfoliative toxin genes, which contribute to the scalded-skin syndrome in humans, have been found in S. agnetis and S. chromogenes strains from poultry (Ladhani et al., 1999). These findings suggest that NAS species in broiler farms, especially CoVS species, may be a potential health hazard for broilers and a potential source of chicken meat contamination in Korea.
Recently, methicillin resistance in NAS strains, especially methicillin-resistant CoNS strains, has been reported in livestock farms and in foods of animal origin (Mahato et al., 2017). Analysis of the SCCmec types of the six mecA-positive NAS strains revealed that two CoNS (S. epidermidis and S. haemolyticus) from broilers or the surrounding environment carried SCCmec V, which is typically associated with methicillin-resistant staphylococci originating from livestock and companion animals (Back et al., 2020; Lee and Yang, 2020). In contrast to SCCmec V in broiler-associated NAS and S. aureus (Lee et al., 2022) strains, the two strains of S. epidermidis from farm workers harbored SCCmec II or SCCmec IV, which has been predominantly found in clinical S. epidermidis isolates from human patients (Barbier et al., 2010). Similar to previous studies, which reported heterogeneity of SCCmec elements in methicillin-resistant NAS isolates (Rolo et al., 2017), SCCmec types of the two mecA-positive CoNS strains, one S. sciuri and one S. ureilyticus strain, could not be determined because of non-typeable ccr/mec genes. Although the prevalence of SCCmec elements in NAS isolates in this study was much less than that of S. aureus in a previously reported study (2.8% vs. 21.5%; Lee et al., 2022) these results indicate that diverse species of NAS may contribute to the transmission of SCCmec elements among staphylococci.
It is estimated that approximately 20% of all antibiotics sold in the livestock industry are used for poultry in Korea (Lim et al., 2014). In particular, β-lactams, fluoroquinolones, and tetracyclines have been extensively used to prevent and treat staphylococcal infections in poultry. Correlating to the administration of antibiotics in poultry, relatively high levels of resistance to AMP, PEN, CIP, and TET were identified in both CoVS and CoNS strains (Figs. 2A and B), indicating that the administration of antibiotics facilitated the occurrence of AMR, including the NAS strains in this study. Although FUS has not been listed as a major antibiotic used in poultry farms in Korea, the highest levels of resistance have been identified in several species of CoVS and CoNS, particularly in S. agnetis (90.2%), S. xylosus (79.6%), and S. saprophyticus (95.0%; Table 2). Previously, it was reported that certain species of NAS, such as S. agnetis, S. saprophyticus, S. cohnii, and S. ureilyticus, frequently carry fusD and fusF for intrinsic resistance to FUS (Chen et al., 2015; O’Neill et al., 2007). The frequent occurrence of antimicrobial-resistant NAS strains, especially MDR strains displaying resistance phenotypes to the antibiotics commonly used in poultry farms, indicates that NAS strains from poultry farms can serve as important reservoirs for AMR.
Frequent occurrence of quinolone resistance has been reported in LA-MRSA and NAS isolates through point mutations in the QRDRs of the two essential enzymes, DNA gyrase and topoisomerase IV (Takahashi et al., 1998; Takahata et al., 1997). In this study, all 57 fluoroquinolone-resistant NAS strains possessed one or more point mutations in the QRDRs of gyrA, gyrB, parC, or parE genes (Table 3). In line with previous studies in S. aureus and CoNS (Lee et al., 2022; Takahashi et al., 1998), mutations at codon 84 of gyrA (S84L and S84F) and codon 80 of parC (S80L, T80I, S80I, and S80V) were most frequently found in fluoroquinolone-resistant NAS strains. In addition to the S84L mutation, nine different amino acid substitutions were identified in QRDRs of gyrA in CoNS strains. High level of heterogeneity in QRDRs of parC was also observed in CoNS strains, especially S. epidermidis, S. gallinarum, and S. xylosus strains. Although none of the fluoroquinolone-resistant CoVS strains had mutations in gyrB and parE,S. sciuri, S. simulans, and S. xylosus carried mutations in the QRDRs of gyrB and parE (Table 3). While S. sciuri and S. xylosus strains had S567A mutation in gyrB, S. simulans strains had A512R, E490G, and S494T mutations in gyrB. As shown in Table 3, only S. simulans strains carried the T379K mutation in parE. Although the T379K mutation has not been previously described, several other mutations in parE have been found in S. aureus (Fujimoto-Nakamura et al., 2005) and Streptococcus pneumoniae (Jones et al., 2000) in association with quinolone resistance. Further studies are required to determine whether the newly identified mutations in gyrB and parE can act synergistically with gyrA and parC mutations to further enhance quinolone resistance in CoNS strains.
In conclusion, our results suggest that (1) there is a relatively high degree of species diversity in NAS isolates, especially CoNS isolates, collected from broiler farms; (2) frequent occurrence of MDR in NAS isolates with high resistance to β-lactams, fluoroquinolones, and fusidane was observed; and (3) fluoroquinolone resistance in broiler-associated NAS isolates was mainly caused by various mutations in the QRDRs of gyrA and parC.













