Introduction
Staphylococci are commensal bacteria that colonize on the skin and mucous membranes of humans and animals (Becker et al., 2014; Casey et al., 2007). However, they are occasionally implicated in local and systemic infections such as scalded skin syndrome, gastroenteritis, and toxic shock syndrome (Ladhani et al., 2004; Lowy, 1998). Although Staphylococcus aureus is most frequently associated with disease outbreaks, recent studies have revealed that non-aureus staphylococci (NAS) substantially affect human and animal health (Adkins et al., 2018; Osman et al., 2017; Wuytack et al., 2020). The consumption or close contact with raw or undercooked meat and other food products contaminated with bacteria are the most common transmission routes from livestock to humans (Osman et al., 2016; Osman et al., 2017; Podkowik et al., 2012). As a zoonotic bacterial pathogen, S. aureus is characterized by the (i) production of coagulase, which converts fibrinogen to fibrin, and (ii) secretion of toxic shock syndrome toxin-1 (TSST-1) and staphylococcal enterotoxins (SEs), which cause staphylococcal food poisoning (SFP; Argudín et al., 2010; Dellaripa, 2000). Although some NAS strains, particularly coagulase-negative staphylococci (CoNS), have one or more genes encoding various SEs, their pathophysiological roles in SFP remain unclear (Podkowik et al., 2013; Wiśniewski et al., 2023). However, antimicrobial resistance (AMR) genes in NAS can be horizontally transferred to confer AMR phenotype in other staphylococci. Notably, studies have revealed a high prevalence of methicillin-resistant S. aureus and MR-NAS in livestock farms, slaughterhouses, and retail meat (Huber et al., 2011; Lim et al., 2010; Schnitt et al., 2021; Van Cleef et al., 2010). The mecA-containing staphylococcal cassette chromosome mec (SCCmec) and other mobile genetic elements (MGEs) carrying AMR genes can be transferred between S. aureus and NAS, which normally co-colonize in livestock such as cattle, pigs, goats, sheep, and poultry (Bhargava and Zhang, 2012; Pyzik et al., 2019; Ray et al., 2016).
Poultry carcasses have been associated with various foodborne pathogens such as Salmonella spp., Campylobacter spp., and Staphylococcus aureus (Creţu et al., 2011; Creţu et al., 2012; Creţu et al., 2015). Moreover, poultry is one of the principal reservoirs for antimicrobial-resistant staphylococci owing to the excessive use of antibiotics in poultry meat production (Apata, 2009; Diarra and Malouin, 2014). High fluoroquinolone (FQ) resistance in chicken-associated staphylococci has led to therapeutic dilemmas in both human and veterinary medicine (Dalhoff, 2012). Although the AMR profiles of S. aureus isolates from poultry and retail chicken meat are annually monitored in several countries including Korea, the USA, and the European Union (Abdalrahman et al., 2015; Feßler et al., 2011; Lee et al., 2022; Normanno et al., 2007), the AMR data for NAS isolates are relatively limited. Several previous studies have revealed various NAS species with AMR, including Staphylococcus gallinarum, Staphylococcus xylosus, Staphylococcus simulans, Staphylococcus arlettae, Staphylococcus chromogenes, Staphylococcus epidermidis, Staphylococcus hyicus, and Staphylococcus lentus in the poultry food chain (Osman et al., 2016; Pimenta et al., 2021; Pyzik et al., 2019). In addition to AMR, most genes encoding SEs located on MGEs can be transferred between NAS and S. aureus, thereby increasing the morbidity and mortality rates of staphylococci (Alibayov et al., 2014).
Previously, we reported the AMR and SE profiles of NAS isolates from healthy broilers (Park et al., 2023) and retail chicken meat (Lee et al., 2020) in Korea. However, NAS species distribution in poultry slaughterhouses and their AMR profiles remain unreported. Therefore, in the present study, we analyzed the species prevalence, AMR phenotypes, and SE gene distribution of NAS isolates obtained from poultry slaughterhouses, including chicken carcasses and facility environments. Furthermore, the major genetic factors associated with methicillin and FQ resistance phenotypes were examined using SCCmec typing and quinolone-resistance determining region (QRDR) sequencing.
Materials and Methods
In total, 270 swab samples were collected from six poultry slaughterhouses located in six Korean provinces from March to December 2019. Swab samples were obtained from chicken carcasses (n=240) within 8 h of slaughter before a chilling process; and slaughterhouse environments (n=30), including cutting boards, sewage, floors, and tables. All swabs were kept at 4°C and delivered to the laboratory within 24 h of sample collection to isolate staphylococci.
For NAS isolation, swabs collected were inoculated into 3 mL of tryptic soy broth (Difco Laboratories, Detroit, MI, USA) supplemented with 10% sodium chloride (TSB-NaCl) and cultured at 37°C for 18–24 h. Next, 10-μL aliquots of pre-enriched TSB-NaCl cultures were streaked onto Baird-Parker agar (Difco Laboratories) containing egg yolk supplements and potassium tellurite. After 24–48 h incubation at 37°C, presumptive staphylococcal colonies were picked from each agar plate and re-streaked on Baird-Parker agar for subsequent identification. Individual isolates were subcultured on tryptic soy agar (Difco Laboratories) at 37°C for 18 h to extract genomic DNA using a Genmed DNA kit (Seoul, Korea) based on the manufacturer’s protocols. NAS species were identified using 16S rRNA sequencing and matrix-assisted laser desorption ionization time-of-flight mass spectrometry (Bruker Daltonics, Bremen, Germany).
All staphylococcal isolates exhibiting cefoxitin resistance were screened for the presence of the mecA gene using polymerase chain reaction (PCR), as described previously (Geha et al., 1994). For mecA-positive isolates, SCCmec types were determined using multiplex PCR, which amplified the chromosomal cassette recombinase genes and mec regulatory elements (Kondo et al., 2007).
To determine the antimicrobial susceptibility phenotype of each NAS isolate, the standard disc diffusion method was used according to the Clinical and Laboratory Standards Institute’s (CLSI) and CLSI VET01S guidelines (CLSI, 2015; CLSI, 2022). Fourteen different antibiotic discs were utilized for the disc diffusion assay on Mueller-Hinton agar (Difco Laboratories): cefoxitin (FOX, 30 μg), penicillin (PEN, 10 μg), ampicillin (AMP, 10 μg), gentamicin (GEN, 50 μg), ciprofloxacin (CIP, 5 μg), chloramphenicol (CHL, 30 μg), erythromycin (ERY, 15 μg), fusidic acid (FUS, 50 μg), clindamycin (CLI, 2 μg), mupirocin (MUP, 200 μg), rifampicin (RIF, 5 μg), sulfamethoxazole-trimethoprim (SXT, 23.75–1.25 μg), tetracycline (TET, 30 μg), and quinupristin-dalfopristin (SYN, 15 μg).
Susceptibilities to vancomycin (VAN), linezolid (LZD), tigecycline (TGC), and teicoplanin (TEC) were examined using a standard Etest (bioMérieux, Craponne, France). Two reference strains, Staphylococcus aureus ATCC 29213 and S. aureus MW2, were included for the disc diffusion assay and Etest.
FQ-resistant NAS isolates frequently carry point mutations within the QRDRs of DNA gyrase (gyrA and gyrB) and topoisomerase IV (parC and parE; Ito et al., 1994; Ng et al., 1996). Genomic DNA from each FQ-resistant isolate was subjected to PCR amplification using QRDR-specific primer sets (Supplementary Table S1). The QRDRs-specific primer sets were designed based on the following published sequences of reference genomes: Staphylococcus agnetis (NCTC7887, NCBI GenBank accession number UHAH01000002.1), Staphylococcus chromogenes (17A, NCBI GenBank accession number NZ_CP031274.1), S. arlettae (P2, NCBI GenBank accession number AP019698.1), S. lentus (H29, NCBI GenBank accession number CP059679.1), Staphylococcus nepalensis (JS1, NCBI GenBank accession number NZ_CP017460.1), S. simulans (MR2, NCBI GenBank accession number NZ_CP016157.1). Then, the PCR products were sequenced at Bionics (Seoul, Korea). Differences in the amino acid sequences of the QRDRs were determined using the nucleotide sequences of the PCR results. Finally, multiple sequence alignments of gyrA, gyrB, parC, and parE genes were analyzed using the CLUSTALW server (www.genome.jp/tools-bin/clustalw).
Multiplex PCR was performed as previously described (Omoe et al., 2005) to examine the carriage of the toxic shock syndrome toxin-1 gene (tst1) and five classical SE genes (sea, seb, sec, sed, and see) in the NAS isolates. Five S. aureus reference strains (N315, MW2, COL, FRI472, and FRI913) were used as positive controls for the SE gene detection.
Results
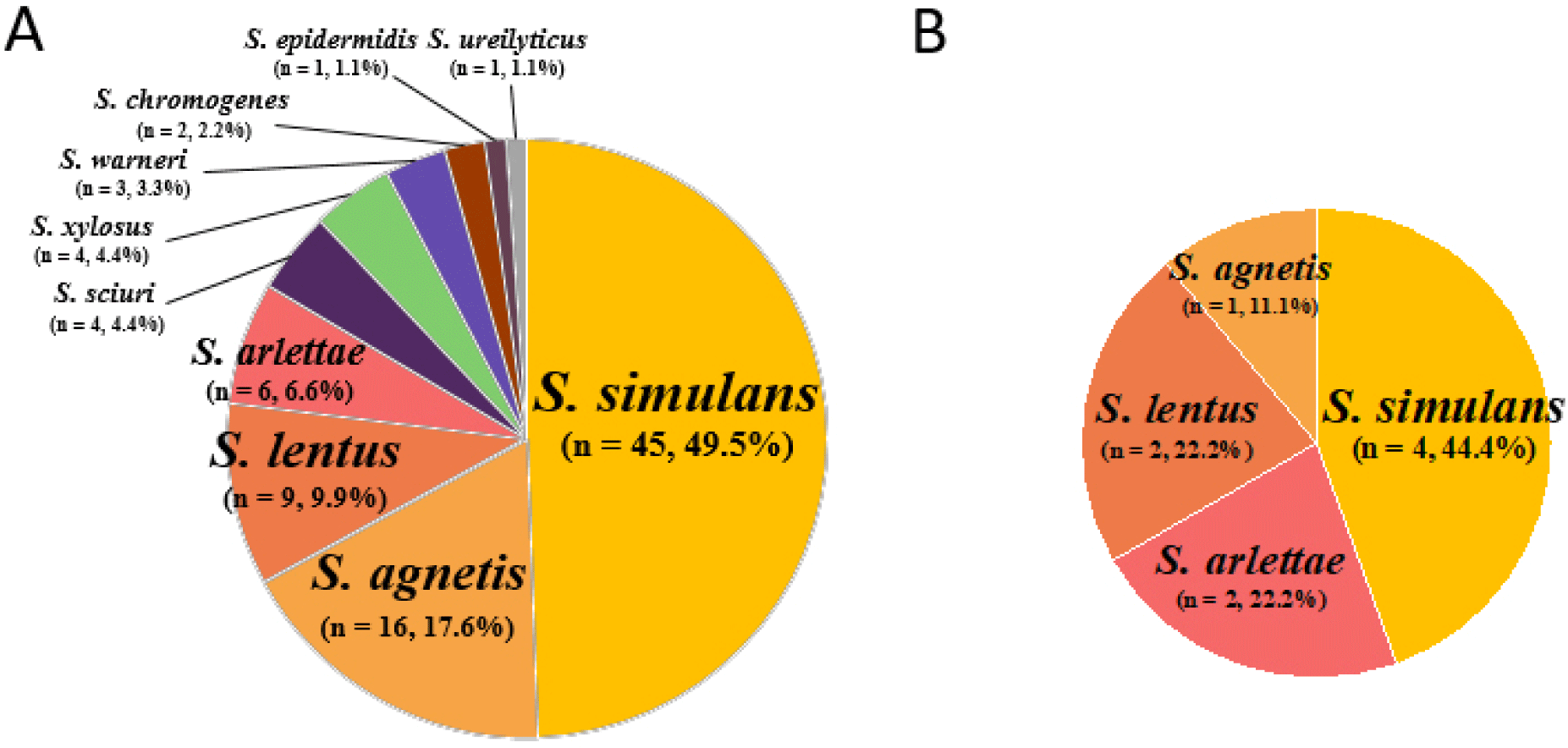
During the study period, 100 NAS isolates of 10 different species were obtained from 270 swab samples (37.04%, 100/270) collected from six different poultry slaughterhouses (Fig. 1A and B). Out of the 100 NAS isolates, 91 NAS isolates were obtained from chicken carcasses, and nine were isolated from the slaughterhouse environments. Based on their ability to produce coagulase, these NAS isolates were divided into coagulase-variable (CoVS; n=19 isolates) and CoNS (n=81; Table 1). While the 19 CoVS isolates comprised only two species of NAS, S. agnetis (n=17) and S. chromogenes (n=2), the 81 CoNS isolates had eight different Staphylococcus species: S. simulans (n=49), S. lentus (n=11), S. arlettae (n=8), S. xylosus (n=4), Staphylococcus sciuri (n=4), Staphylococcus warneri (n=3), S. epidermidis (n=1), and Staphylococcus urealyticus (n=1). The two most prevalent NAS species isolated from the chicken carcasses were S. simulans (49.5%, 45/91 isolates) and S. agnetis (17.6%, 17/91 isolates; Fig. 1A). Similarly, the nine NAS isolates from the facility environments were S. simulans (n=4 isolates), S. arlettae (n=2), S. lentus (n=2), and S. agnetis (n=1; Fig. 1B).
PCR analysis revealed that none of the CoVS isolates were positive for mecA gene (Table 1). Among the 81 CoNS isolates, 15 (18.5%) were positive for mecA: S. simulans (n=8), S. lentus (n=6), and S. epidermidis (n=1). Furthermore, SCCmec element typing of the 15 mecA-positive NAS isolates revealed that eight S. simulans isolates had either non-typeable SCCmec (n=6) or SCCmec I (n=2). In addition, all six methicillin-resistant S. lentus strains contained non-typeable SCCmec elements. One methicillin-resistant S. epidermis isolate from a chicken carcass sample possessed SCCmec IV.
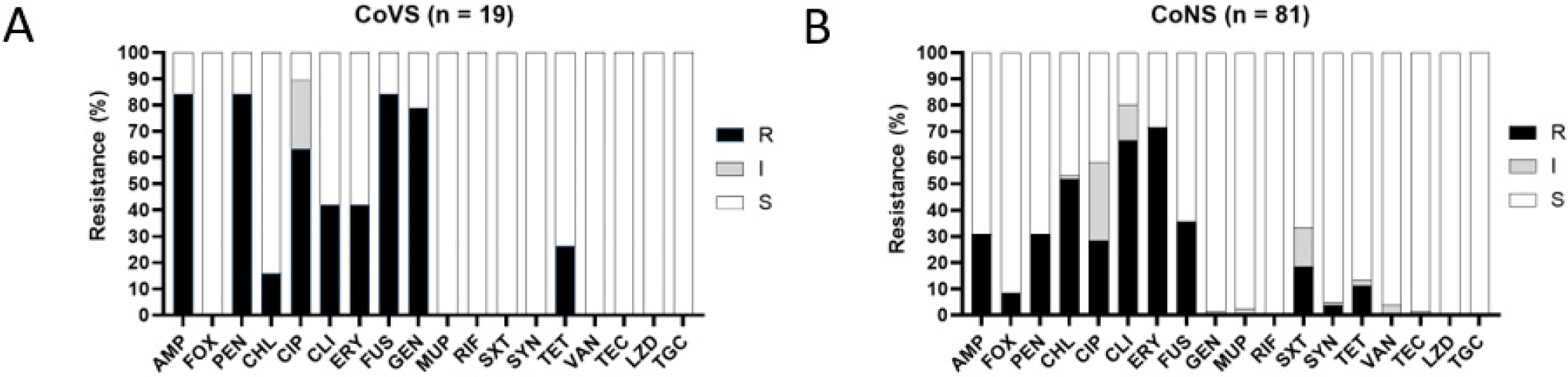
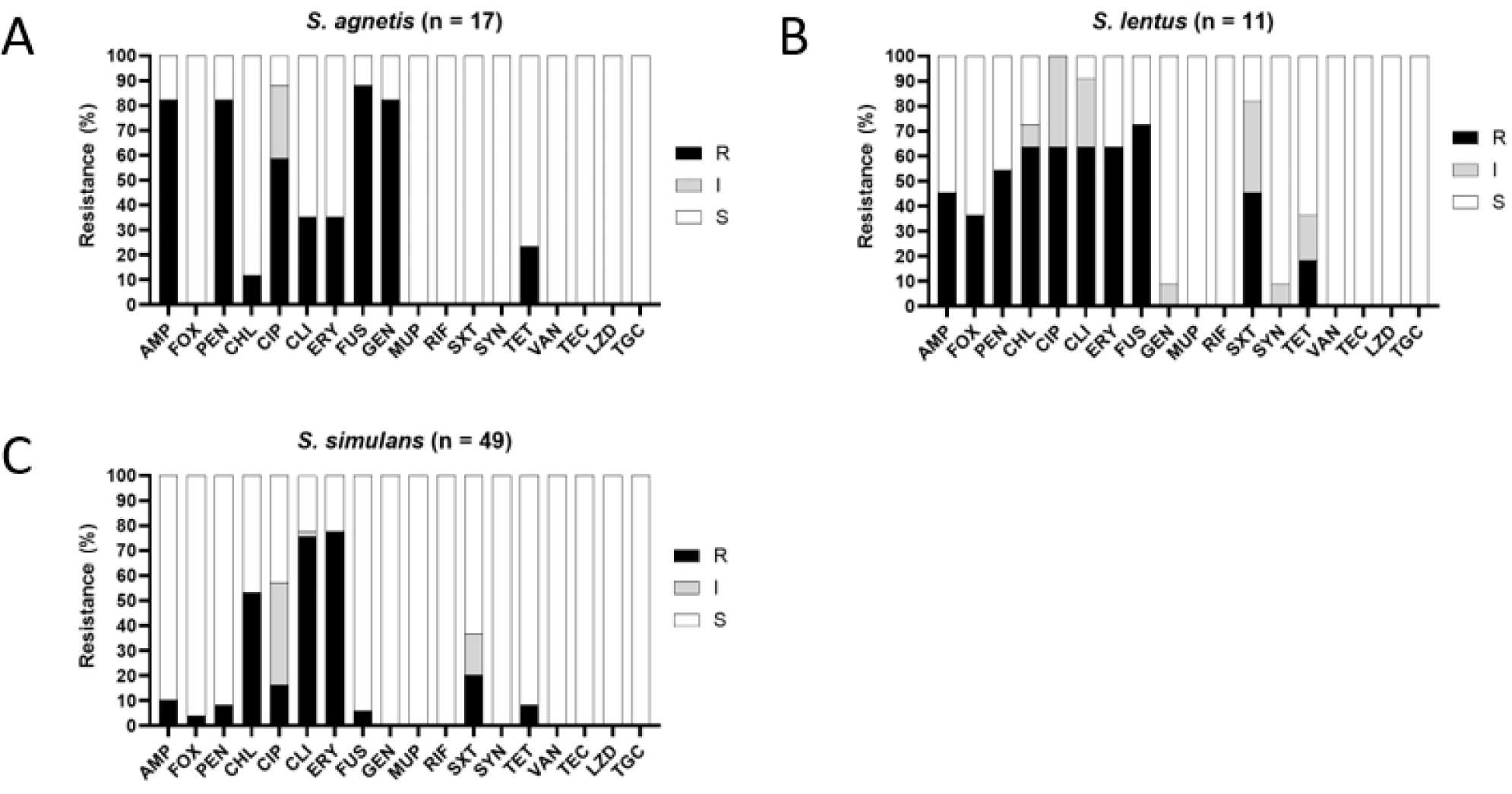
Compared with the other antibiotics, the CoVS isolates exhibited higher resistance to FUS (84.2%, 16/19 isolates), PEN (84.2%, 16/19 isolates), AMP (84.2%, 16/19), GEN (78.9%, 15/19), and CIP (63.2%, 12/19; Table 2 and Fig. 2). Although a higher resistance level to these antibiotics was also observed in CoNS isolates, CoNS exhibited lower resistance rates to the five antibiotics compared with CoVS isolates. In addition, CoVS isolates exhibited higher levels of multidrug resistance (MDR) phenotypes (resistant to three or more classes/subclasses of antimicrobial agents) than CoNS isolates (84.2% vs. 63.0%, respectively). When the AMR of the dominant CoVS species (S. agnetis) and the two major species of CoNS (S. simulans and S. lentus) were compared, S. agnetis (82.4%) and S. lentus (90.9%) exhibited significantly higher degrees of MDR than S. simulans (57.1%; Table 2). Fig. 3 illustrates the heterogeneity of the AMR profiles within the three major species of NAS isolates, S. simulans, S. agnetis, and S. lentus. S. agnetis isolates tended to show high resistance to AMP, GEN, PEN, CIP, and FUS (Fig. 3A). Interestingly, S. lentus, and S. simulans isolates exhibited rather distinct AMR profiles (Fig. 3B and C). However, both S. lentus and S. simulans displayed comparatively high levels of resistance to CHL, CIP, CLI, and ERY. Notably, S. agnetis and S. lentus isolates exhibited higher levels of CIP resistance (61.1% and 63.6%, respectively) than S. simulans isolates. Antimicrobial resistance profiles of each strain are shown in Supplementary Table S2.
NAS, non-aureus staphylococci; AMP, ampicillin; FOX, cefoxitin; PEN, penicillin; CHL, chloramphenicol; CIP, ciprofloxacin; CLI, clindamycin; ERY, erythromycin; FUS, fusidic acid; GEN, gentamycin; MUP, mupirocin; RIF, rifampin; SXT, trimethoprim-sulfamethoxazole; SYN, quinupristin- dalfopristin; TET, tetracycline; MDR, multi-drug resistance; CoVS, coagulase-variable staphylococci; CoNS, coagulase-negative staphylococci.
Among the 100 NAS isolates, 12 CoVS (10 S. agnetis and two S. chromogenes) and 22 CoNS (eight S. simulans, seven S. arlettae, and seven S. lentus) isolates exhibited resistance to CIP (Table 3). Sequencing analysis of QRDRs within gyrA, gyrB, parC, and parE revealed that mutations at codon 84 in gyrA (S84L and D84N) and codon 80 in parC (S80L, T80I, T80R, S80F, and S80Y) were most frequently associated with the CIP-resistant phenotype of the NAS isolates (Table 3). Unlike point mutations in gyrA and parC, a high degree of mutation heterogeneity was observed in gyrB and parE within and between different NAS species (Table 3). Nevertheless, none of the FQ-resistant S. lentus (n=7) isolates carried point mutations in gyrB. Similarly, none of the FQ-resistant S. agnetis isolates harbored point mutations in parE.
Among the 100 NAS isolates, only two CoNS isolates (one S. simulans and one S. xylosus) from chicken carcasses were positive for TSST-1 gene. None of the 19 CoVS isolates were positive for the five classical SE genes and tst1.
Discussion
Many recent studies have revealed that NAS are important reservoirs of AMR and enterotoxin genes, which can be transmitted to other pathogenic bacteria, including more pathogenic strains of S. aureus. In our previous research, we suggested that NAS in retail chicken meat can play an important role in the transmission of AMR and enterotoxin genes (Lee et al., 2020). However, not many studies have investigated the species profiles of antimicrobial-resistant NAS and their significance in food-related public health.
In the present study, we investigated NAS species distribution and their AMR profiles in chicken carcasses, facility environments, and poultry slaughterhouse workers in Korea. Similar to the findings of our previous studies on NAS profiles in broiler farms (Park et al., 2023) and retail chicken meat (Lee et al., 2020), the prevalence of CoNS (81.0%, 81/100) was considerably higher than that of CoVS (19.0%, 19/100; Table 1). A higher prevalence of CoNS compared with CoVS has also been reported in both healthy broilers and broilers with signs of illness from other countries. In the present study, S. simulans (49.0%, 49/100) and S. agnetis (17.0%, 17/100) were the most frequently identified NAS species in poultry slaughterhouses; this is consistent with the previously reported high prevalence of S. agnetis, S. simulans, S. haemolyticus, and S. xylosus in broiler farms (Park et al., 2023). Sample origin did not affect the species distribution of NAS isolates in chicken carcasses and facility environments in poultry slaughterhouses (Figs. 1A and B). The four major NAS species, S. simulans, S. agnetis, S. lentus, and S. arlettae, were commonly identified in both chicken carcasses and facility environments. These results suggest that broiler-associated NAS species (Park et al., 2023), both CoVS and CoNS species are potential sources of contaminants in chicken carcasses and facility environments in poultry slaughterhouses.
Approximately 20% of all the antibiotics sold in the livestock industry, particularly FQs, β-lactams, and TETs, are consumed in poultry farming in Korea (Lim et al., 2014). Thus, recent studies on livestock-associated antimicrobial-resistant NAS, particularly methicillin-resistant NAS isolates, have raised significant concerns regarding the transmission of resistance genes to more pathogenic bacteria in the food production system. In a previous study in Switzerland (Huber et al., 2011), researchers reported that 52.8% (38/72) of mecA-positive NAS isolates, i.e., S. lentus (n=30), S. sciuri (n=6), and S. epidermidis (n=2), were obtained from chicken carcasses. In the present study, mecA-positive NAS isolates (15.0%, 15/100) were present at relatively low levels in poultry slaughterhouses. As summarized in Table 1, eight mecA-positive but FOX-susceptible NAS isolates (two S. lentus and six S. simulans) carrying non-typeable SCCmec elements were identified. Recent studies revealed the mecA-positive but FOX-susceptible phenotype of staphylococci (Goering et al., 2019; Ho et al., 2020). This discrepancy could be owing to the altered expression of mecA because of dysregulation in mecR1 (encoding the inducer protein MecR1), mecI (encoding the repressor protein MecI), or bla regulatory system (blaR1-blaI; Liu et al., 2016). In addition, a single-nucleotide insertion within mecA, resulting in the premature termination of mecA expression, has been identified in S. aureus strains (Kime et al., 2019).
Largely correlating with the sales amounts of antibiotics in the poultry industry in Korea, relatively high levels of resistance to β-lactams, GEN, TET, and CIP were identified in NAS isolates, particularly in CoVS isolates. This suggests that antibiotic selective pressure affect the prevalence of antimicrobial-resistant NAS isolates in poultry slaughterhouses (Table 2). Furthermore, the high prevalence of FUS resistance in some of NAS isolates, particularly S. agnetis, S. arlettae, S. lentus, and S. xylosus, may be because of intrinsic FUS resistance owing to the frequent carriage of fusD and fusF (Hung et al., 2015). Notably, the high prevalence of NAS isolates with MDR phenotypes to the antibiotics extensively consumed in poultry farms suggests that antibiotic selective pressure facilitates the proliferation of antimicrobial-resistant NAS in broiler farms, and thus, its transmission to chicken carcasses in slaughterhouses.
The World Health Organization has categorized FQs as the highest-priority critically important antibiotics (Scott et al., 2019). However, FQ-resistant S. aureus and NAS strains have been frequently isolated in livestock farms, particularly poultry farms, in Korea. In this study, 34 FQ-resistant NAS isolates (12 CoVS and 22 CoNS) were identified to harbor multiple point mutations in the QRDRs of DNA gyrase and topoisomerase IV, except for one S. arlettae isolate carrying a single point mutation in parC (Table 3). Consistent with the findings of previous studies of FQ-resistant staphylococci (Takahashi et al., 1998; Wang et al., 2013), all 34 FQ-resistant NAS isolates possessed point mutations in gyrA (codon 84) or parC (codon 80). In contrast to the mutations in gyrA and parC, which are frequently concentrated in codons 84 and 80, respectively, point mutations in gyrB and parE were identified at various locations in QRDRs. Moreover, previous studies on FQ-resistant staphylococci (Nakaminami et al., 2014; Yamada et al., 2008) revealed a high degree of heterogeneity in the point mutations in gyrB and parE. Although major mutations, including S84L at gyrA, S80L at parC, and D84N at parC, were previously known to be associated with FQ resistance (Li et al., 1998; Takahashi et al., 1998; Vila et al., 1997), the precise role of other minor mutations in FQ resistance should be elucidated in the future research.
SEs produced by coagulase-positive S. aureus are a major cause of SFP (Argudín et al., 2010; Fisher et al., 2018). However, many recent studies have indicated that NAS isolates, including those from livestock and food products, carry SE genes and are potentially enterotoxigenic (Mekhloufi et al., 2021; Podkowik et al., 2013). In this study, our results demonstrate that CoVS isolates from poultry slaughterhouses do not harbor the classical SE nor tst-1 genes. Although previous studies have revealed the highest prevalence of sec and tst1 in CoNS strains (Hájek, 1978; Valle et al., 1990), only two tst1-positive (one S. simulans and one S. xylosus) NAS isolates were detected in the present study. Since SEA and TSST-1 were first detected in CoNS strains isolated from patients with toxic shock syndromes (Crass and Bergdoll, 1986), potential enterotoxigenic or pathogenic NAS strains carrying SE genes have been detected in humans (Achek et al., 2018; Banaszkiewicz et al., 2022), livestock (Ruiz-Ripa et al., 2020; Ünal and Çinar, 2012), and ready-to-eat foods (Chajęcka-Wierzchowska et al., 2020; Crass and Bergdoll, 1986; Cunha et al., 2006; Rall et al., 2010; Zell et al., 2008). Despite the proposed instability of SE genes in CoNS isolates (Banaszkiewicz et al., 2022), the carriage of SE and TSST-1 genes in NAS isolates from poultry slaughterhouses should be monitored as a significant threat to food safety.
Conclusion
In summary, this is the first report on the species diversity, AMR patterns, and genetic characteristics of methicillin- and FQ-resistant NAS isolates obtained from poultry slaughterhouses in Korea. Our findings suggest that (i) the overall species distribution of NAS isolates in poultry slaughterhouses is similar to that in broiler farms (Park et al., 2023) in Korea; (ii) MDR phenotypes with relatively high resistance levels of resistance to PEN, AMP, GEN, CIP, and TET are observed in NAS isolates; (iii) FQ-resistance in NAS isolates is caused by point mutations occurring at specific locations (gyrA and parC) or rather various locations (gyrB and parE) of QRDRs; and (iv) the carriage of tst1 only in CoNS isolates, indicating the extremely low prevalence of the TSST-1 and SE genes in NAS isolates. In addition, this study underscores a potential role of poultry slaughterhouses in transmission of antimicrobial-resistant NAS strains to human food chains.
To combat AMR in poultry industry, a multisectoral approach, including poultry farms, slaughterhouses, and human workers, is necessary. Improper use of antibiotics and indiscriminate prescription of antibiotics should be avoided to prevent occurrence and amplification of antimicrobial-resistant bacteria in poultry farms. Furthermore, importance of hygiene measures in slaughterhouses should be emphasized along with a multisectoral surveillance networks to better detect and mitigate spread of AMR.













