Introduction
Beef is one of the most popular livestock commodities in Indonesia, which is consumed as meat and processed products such as meatball. Increasing beef demand as a main ingredient of meatballs, which is not balanced with sufficient supply of beef, creates an opportunity to adulterate and mix beef with other meat sources. Consumers find it difficult to prove which original beef or adulterated beef by bare eyes; it is even more challenging to identify beef adulteration in processed products. Adulteration of raw beef with other meats and its processed products can cause various problems, including consumer satisfaction, social problems, particular religious preferences, and other problems related to health hazards associated with certain types of substances from certain types of meat (Kumar et al., 2014).
Cases of beef adulteration are not only limited to substitution with pork (Ha et al., 2017). In Indonesia, beef adulteration is also mixed with dog and rat meats due to both types of meat is served as food in some cities (Weichart, 2004). The level of dog meat consumption in Asia, especially in Indonesia, is reasonably high because of eating dog meat is culturally permitted behavior in several Indonesian regions. It is reported that around one thousand dogs are slaughtered and served as food every week (Podberscek, 2009; Weichart, 2004). In the Europe, the idea of consuming dog meat is seen as something disgusting because, generally, dogs are kept as pets (Podberscek, 2009). Meat from dog, pig, and rat species has been regulated by criteria for prohibited consumption in Islamic law (Fadzlillah et al., 2011). In addition, these types of animals also have the potential to become zoonotic agents (Fajardo et al., 2010; Hamidi, 2018). If this type of meat is used in multi-species adulteration, it will be harmful not only for Moslem but also for all consumers.
Labeling regulation requires that compositions of meat products must be clearly stated in the package to protect the worldwide consumer from deviant meat products (Abuzinadah et al., 2015; Doosti et al., 2014). Stating composition in the package is obviously intended to protect consumers from fraud and adulteration. Sometimes producers of beef product do not provide clear information in the package, and they are often deliberately mixed beef with pork and its derivatives due to the obvious economic benefits in some countries like Korea, Japan, and China where beef is much expensive than pork (Ha et al., 2017).
The DNA-based method much more effective than using physical properties of meat in determining of meat species origin (Tathma et al., 2019). The application of this method for identification species origin contained in meat products using polymerase chain reaction (PCR) technique is frequently applied because DNA is quite stable against high temperature, pressure, and chemical treatments (Fajardo et al., 2010; Saez et al., 2004). The multiplex-PCR method uses more than one primer in one PCR tube. Therefore, it can be used to analyze large sample quantities, saving time, cost, and it is also highly sensitive (Dalmasso et al., 2004). Most of the previous studies develop mitochondrial genome such as Cytochrome b and 12S rRNA genes as target sites for phylogenetic, evolution, and species identification studies due to their unique nucleotides and maternally inherited to the offspring. The 12S rRNA region has been used for species identification in raw meats using multiplex-PCR assay (Cahyadi et al., 2018; Cahyadi et al., 2019). Therefore, the objective of this study was to detect intentionally contaminated beef meatball with three non-halal meats (dog, pork, and rat) using multiplex-PCR with the 12S rRNA region designed as target sites.
Materials and Methods
The samples of beef and pork were collected from traditional markets in Surakarta City. Dog and rat meats were collected from meat sellers who are not willing to be published to the public. Each meat sample was put into a zip locked plastic bag and tagged with a specific name and stored separately to avoid cross-contamination among samples until used for further process in the refrigerator at 4°C.
Meats were thawed and cut into small pieces using sharp knives. They were ground using different meat grinder for each meat species (Meat Mincer LH-22CW, Huamei, Zhejiang, China). Ground meats were gently mixed with wheat flour, garlic powder, salt, and pepper until homogeneous. Ingredient of each sample unit is presented in Table 1. Furthermore, meatball doughs were shaped like a ball by hand, and then they were boiled for 20 minutes until floating. Cooked meatballs were drained and left at room temperature. Finally, they were separately stored in the freezer at −20°C until used for the subsequent analysis. The process of making meatball in this study used different equipment for each sample unit to prevent cross-contamination among samples (Sari et al., 2017).
Meatball samples in this study were divided into eleven sample units (Table 1). Every unit has three replications. Samples of S, A, B, and T meatballs were set for simplex-PCR, duplex- (SA, SB, and ST), and multiplex-PCR (SAB, SAT, SBT, and SABT) were made based on a combination of the composition of meat from beef (S), dog (A), pork (B), and rat (T). Those samples were used to test the uniqueness and specificity of the 12S rRNA gene primers using genomic DNA templates extracted from meatballs.
The genomic DNA was extracted according to the procedure of gsyncTM DNA extraction kit for tissue (Geneaid Biotech, Taipei, Taiwan). The isolated genomic DNA was stored at −20°C until used for PCR (Yacoub and Sadek, 2016). To evaluate the quality of isolated genomic DNA, 1% agarose gel electrophoresis was performed at 100 Volts for 30 minutes. Moreover, stained agorose gel was put into gel document to capture photograph (Bio-Rad Gel Doc™ XR +, USA).
The genomic DNA extracted from the meatball sample was used as a DNA template for the purpose of obtaining specific segments of the 12S rRNA gene as target using primers previously reported by Cahyadi et al. (2018) and Cahyadi et al. (2019). Primer pairs are shown in Table 2. Simplex-, duplex-, and multiplex-PCR reactions were carried out using a thermal cycler machine (GeneAmp® PCR System 9700, Singapore). The total volume of reaction in the microtube was 25 μL consisting of 12.5 μL MyTaq™ HS Red Mix (Bioline, London, UK), 1 μL genomic DNA template (10 ng/μL), 1 μL forward primer, 1 μL each reverse primer, and ddH2O adjusted until reach 25 μL total volume. Simplex-PCR only contained one primer pair, duplex-PCR contained two primer pairs (bovine primers and another species primers), and multiplex-PCR contained all primer pairs. The concentration of each primer in the reaction was 10 μM. The PCR reaction was started with an initial denaturation at 95°C for 3 minutes and followed by 30 cycles of denaturation at 95°C for 15 seconds, annealing at 64°C for 30 seconds, and then extension at 72°C for 30 seconds. The PCR process was completed by final extension at 72°C for 3 minutes. Finally, PCR products were electrophoresed using 2% agarose gel at 100 Volts for 30 minutes. Electrophoresis results were visualized using a gel document machine (Gel Doc™ XR+, Bio-Rad, Hercules, CA, USA). The 100 bp marker ladder was used as a standard size of the DNA band.
Results
Simplex-PCR was conducted using meatball samples containing one species of meat origin. The result showed that primer pairs specifically for bovine, dog, pig, and rat correctly amplified target sites. Physical and heat treatment of samples during meatball-making did not affect DNA amplification. Simplex-PCR was successfully performed which is indicated by 155, 244, 357, and 491 bp DNA bands for bovine, dog, pig, and rat, respectively (Fig. 1). The simplex-PCR products were also evident. It indicated that DNA extracted in this study was pure, and no other DNA sources originated from other ingredients or RNA contamination. Therefore, every component in the reaction can be simultaneously working to produce specific and clear DNA bands.
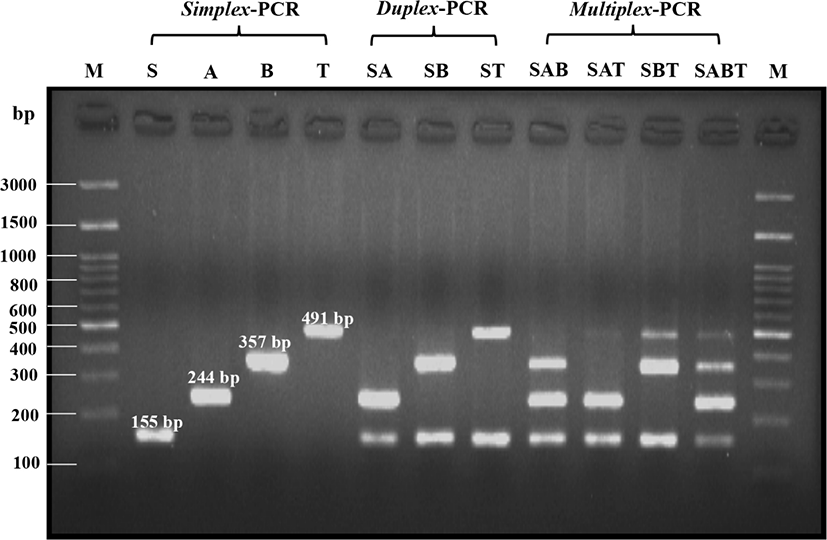
The aim of duplex-PCR was to check the possibility of non-halal meats contaminating beef. The beef was intentionally contaminated with pork, dog, and rat meats that represented non-halal meats. The result showed that duplex-PCR was also successfully carried out. Two DNA bands can be noticed corresponding to bovine and dog/porcine/rat specific segments (Fig. 1). The DNA bands were also clear and bright. These results suggested that two DNA genomics successfully extracted from treated meats with excellent purity and concentration of DNA genomes.
Triplex- and tetraplex PCRs represented multiplex-PCR. In these tests, beef samples were mixed all together with other meats. The existence of three DNA bands indicated the triplex PCR. Also, the tetraplex PCR was indicated by four DNA bands identified in the agarose gel. The results of multiplex-PCR using the 12S rRNA gene as target sites indicated that this novel multiplex-PCR was powerful enough to identify non-halal meats in meatballs. The DNA bands can be seen in the samples containing multi DNA sources, except the SAT sample (Fig. 1). The DNA band for the rat in the SAT mixture was slight, and it was not as bright as other bands. It may be due to the low concentration of rat genomic DNA extracted from the SAT sample; however, it still can be seen by the bare eyes. To prove specificity of primers, multiple alignment analysis of the 12S rRNA gene sequences obtained from genbank is provided in the Fig. 2.

Discussion
The DNA bands of each species in the simplex-, duplex-, and multiplex-PCR were explicitly detected. The primer pairs accurately amplified target sites of the 12S rRNA gene. Heat and physical treatment of meats did not affect the PCR results. The reactions were working well, which are indicated by specific and unique DNA bands for each species. The previous study reported that high processing temperatures up to 160°C did not affect the stability of the mitochondrial DNA 12S rRNA gene. It still can be a target of amplification in the PCR (Lakzadeh et al., 2013). The primer pairs of the 12S rRNA gene used in the present study can amplify the target area of the 12S rRNA gene using genomic DNA extracted from meatball samples. This result was in agreement with previous studies that successfully detected non-halal meats in raw meats (Cahyadi et al., 2018; Cahyadi et al., 2019).
Detection of a mixture of beef, dog, pig, and rat in raw meat and processed products such as meatballs has also been developed previously using the Cyt b gene (Novianty et al., 2016; Primasari, 2011; Rahman et al, 2014). The 12S rRNA gene primer in the present study has advantages, namely using universal forward primers and specific reverse primers to produce specific results. The primary 12S rRNA gene also has the same annealing temperature, so the multiplex-PCR, which is using more than one primer pairs in one tube reaction, generates more accurate amplification targets (Cahyadi et al., 2019). The PCR product size for each species was consistent in the simplex-, duplex-, and also multiplex-PCR. In the SAT meatball, the DNA band of rat species was less bright than other samples containing rat meat. It may be due to a low concentration of extracted rat DNA genomic. Also, the DNA concentration obtained from mix samples cannot be equal even though the meat proportion in the SAT samples was identical. Every part of the meatball cannot be extracted entirely since the reaction during the DNA extraction process cannot be adequately controlled by hand. Degradation of DNA molecules due to heat and physical treatments during processing may also lead to generating low DNA concentration and amplicon of PCR (Di Pinto et al., 2007). The less bright DNA band of multiplex-PCR samples may also be due to unspecific and interference among primers. However, the primer designed in this study was carefully checked by multiple alignment analysis of 12S rRNA gene sequences. The results suggested that reverse primers were specific for each species. It was proved by no identical nucleotide(s) at the three-prime end (3’-end) found in reverse primers. This criterion is absolutely necessary since different nucleotide(s) at the 3’-end of the primer is required for successful multiplex-PCR (Matsunaga et al., 1999).
Utilization of the 12S rRNA gene for species identification is previously reported that it can identify species of origin in the feedstuffs up to 0.01% of the DNA genome containing in the sample (Safdar and Junejo, 2015). In terms of using meatballs, earlier reports explained that duplex-PCR could detect up to only 1% chicken and pork contamination in the meatball made from beef (Novianty et al., 2016; Sari et al., 2017).
This study developed the mitochondrial DNA 12S rRNA gene as a biomarker for non-halal meats detection in processed meat product. This study also focused on identifying more than one species prohibited to be eaten by Moslem using the 12S rRNA gene by developing a multiplex PCR technique. There is no report yet regarded using the multiplex-PCR 12S rRNA gene to identify species in the animal-based product. Therefore, it could be a handy tool to be applied in the vast areas of the food industry. The multiplex-PCR can simultaneously amplify several different DNA sequences and gain more information in a single reaction, which leads to more effective and efficient using conventional PCR. On the other hand, the development of other PCRs such as PCR, PCR-RAPD, PCR-RFLP, species-specific PCR, and real-time PCR to detect species in food products are considered to be less efficient because they require the latest technology and high operational costs than multiplex-PCR (Fajardo et al., 2010). The main motive for most beef adulteration cases revealed in Indonesia is by mixing beef with large quantities of other animal meats to reduce production costs and to get more financial benefits (Ha et al., 2017). The enactment of Law No. 33 of 2014 concerning Guaranteed halal products in Indonesia makes the method of detecting non-halal ingredients in meat-based foods essential.
This novel multiplex-PCR with the 12S rRNA gene as a target for amplification could be promising tools to detect existences of porcine, rat, and dog in meat products in supporting Indonesian government policy. This novel finding could be an alternative DNA-based testing method to identify non-halal meats and their derivative materials in foods.













