Introduction
Ezine cheese is a well-known Turkish cheese that is produced at large scales and is consumed in great quantities in Turkey. With its protected designation of origin (PDO) status, Ezine cheese is also registered by Turkish authorities as a unique trademark. In the northwestern region of Turkey, Ezine cheese is primarily produced in the towns of Ezine, Ayvacık, and Bayramiç. In this region, Ezine cheese is produced from a mixture of pasteurized ewe’s milk (approximately 45%–55%), goat milk (at least 40%) and cow milk (the most 15%) from March to July (Anonymous, 2006). Although Ezine cheese is classified as a white-brined cheese, it is different from classic Turkish white-brined cheeses in that its production involves the use of a mixture of milk and does not utilize a starter culture or CaCl2 and has long ripening periods (TS, 1995).
It is well known that a wide variety of microbiotas cause many microbial and biochemical changes during the ripening period of cheese (Fuquay et al., 2011; Gobbetti et al., 2015; Montel et al., 2014; Pappa et al., 2016). In particular, lactic acid bacteria (LAB) play a significant role in the development of the flavor, texture, and nutritional value of cheese by producing acids and several lipolytic, proteolytic, and glycolytic enzymes. In addition to these crucial roles, the antimicrobial properties of the LAB, such as the production of organic acids, hydrogen peroxide and antimicrobial peptides (known as bacteriocins), reduce the risk of the growth or survival of foodborne pathogens and spoilage organisms, resulting in the improved microbiological stability and extended shelf life of cheeses (Montel et al., 2014; Settanni et al., 2010). Similarly, non-starter lactic acid bacteria (NSLAB), which originate from the raw milk, cheese-producing environment, airborne flora or as a result of unsuccessful pasteurization have substantial effects on the ripening process and the creation of the final sensorial characteristics of cheese (Fuquay et al., 2011; Gobetti et al., 2015; Montel et al., 2014). NSLAB, mainly mesophilic lactobacilli, leuconostocs, enterococci, and pediococci, allow traditional cheeses to develop their particular characteristics, such as low pathogen risk and diversification of gustatory characteristics. NSLAB rapidly reach high counts and act as an acidifying starter that leads to regulate the growth of the cheese microbiota (Montel et al., 2014).
In recent years, there has been a growing interest in isolating the naturally occurring microbiota of cheeses, primarily from those produced without the addition of starter cultures. Furthermore, due to the low variety of starter cultures used in the dairy industry, cheesemakers desire the isolation of novel LAB strains used as indigenous starters (Bozoudi et al., 2016; Dolci et al., 2008). Traditional cheeses produced without the addition of starter cultures, such as Ezine cheese, have a high potential for the isolation of new strains that can be used in the dairy industry. Moreover, as a result of these studies, the selection of strains with technological value and that have the ability to produce bacteriocins against pathogenic or spoiling microorganisms may have significant applications for the large-scale production of traditional cheeses (Gonzales et al., 2007). Although some aspects of the sensory characteristics and aroma compounds of Ezine cheese are known (Yuceer et al., 2009), the chemical and microbiological characteristics of Ezine cheese have not been reported to date. Thus, the objectives of this study were i) to follow the changes in the physicochemical composition and the microbial evolution during the ripening period, ii) to isolate strains with antimicrobial activity that can be used as a starter or bacteriocin producer culture to produce Ezine cheese, and iii) to identify NSLAB strains by the phenotypic and genotypic methods. To the best of our knowledge, this is the first detailed study that explores the chemical and microbiological characteristics of Ezine cheese, the results of which have the potential to guide Ezine cheese producers for the large-scale production of cheese using starter/adjunct cultures.
Materials and Methods
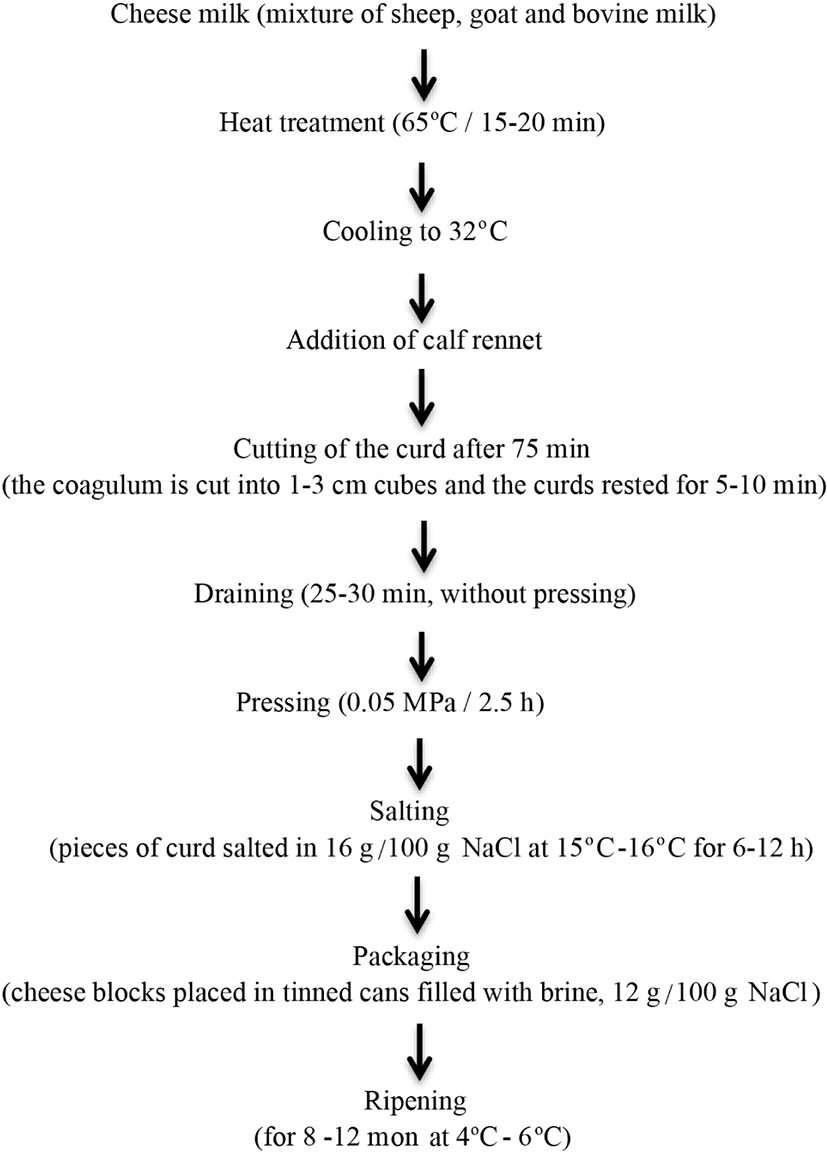
The samples were collected from randomly chosen dairies that follow traditional manufacturing procedures (Fig. 1) located in Ezine, Ayvacık, and Bayramiç, which were indicated as PDO. The study samples were transferred in to the tin plate containing brine and were ripened for 12 mon at the dairies’ temperature-controlled cellars (4°C–6°C). The samples ripened in dairies E, A, and B at the same conditions. During the ripening period, totally 144 cheese samples from three dairy farms were transferred to the laboratory on ice, and chemical and microbiological analyses were subsequently performed.
For the identification process, the letters E, A, and B were used to refer the samples collected from the dairies in Ezine, Ayvacık, and Bayramiç, respectively. The letters a, b, c, and d were used to denote the cheeses produced in April, May, June, and July, respectively, and the numbers 1 to 12 were used to represent the ripening months.
According to the International Dairy Federation (IDF) standards, the percentages of fat (IDF, 2008), total solids (IDF, 1982), lactic acid (IDF, 2005), and NaCl (IDF, 1988) in samples were determined. Dry matter (DM) (IDF, 1986) was determined by weighing the samples before and after the drying process at 102±1°C. The pH values of the samples were measured using a pH meter (Sartorius, Goettingen, Germany).
Ten grams of each cheese sample was transferred to 90 mL of sterile quarter-strength Ringer’s solution (Merck). Cheese samples were homogenized in a stomacher (AES Chemunex, Easy mix) for 2 minutes, and serial dilutions were prepared by mixing 1 mL of the homogenized sample with 9 mL of sterile quarter-strength Ringer’s solution. NSLAB were enumerated and identified by plating samples on M17 Agar (Merck) and DeMan, Rogosa and Sharpe (MRS, Merck) Agar containing 20 μg mL−1 vancomycin (V, SigmaTM) to isolate lactococci and leuconostocs, respectively, after aerobic incubation at 30°C for 72 h. Rogosa Agar (Merck; anaerobic incubation at 30°C for five days) and Kanamycin Aesculin Azid Agar (KAA) (Merck; aerobic incubation at 37°C for two days) were used to enumerate and isolate lactobacilli and enterococci, respectively (Navidghasemizad et al., 2009). Total mesophilic aerobic bacteria (TMAB) and yeast and molds (YM) were counted by plating serial dilutions on Plate Count Agar (PCA) (Merck; aerobic incubation at 30°C for two days) and Yeast Extract Glucose Chloramphenicol Agar (YGEC) (Merck; aerobic incubation at 25°C–28°C for five days), respectively. For each microbial group, the experiments were repeated in triplicate. Following the incubation, the plates with 10–300 colonies were selected for enumeration, and the number of colonies was expressed as Log CFU/g.
To isolate NSLAB with antimicrobial activity, agar spot tests were performed. Briefly, the colonies were randomly selected from each medium and spotted onto M17, MRS, and Brain Heart Infusion Agar (BHI, Merck) plates using sterile toothpicks. After overnight incubation, 5 mL of soft agar (0.7% agar-agar) containing 106 CFU/mL of the indicator strains (Lactococcus lactis ssp. lactis SIK83, Lactobacillus sakei NCDO 2714 and Enterococcus faecalis ATCC 29212) was poured over plates. The plates were incubated overnight at suitable temperatures according to the optimum growth temperature for the indicator strains, and the colonies subsequently were examined (Schillinger and Lucke, 1989). Colonies with clear inhibition zones (≥1 mm) were purified using same media and then were stored in M17, MRS and BHI broth containing 20% (v/v) glycerol at −20°C. According to the diameter of inhibition zones, the level of the antimicrobial activity of the isolates was evaluated as + (1–10 mm), ++ (11–20 mm), +++ (≥21 mm). The working cultures were prepared from frozen cultures by transferring cells into the appropriate broth and incubating overnight.
Isolates from M17 and Rogosa agar plates were examined for Gram staining characteristics, catalase reaction, CO2 production from glucose, arginine hydrolysis, and growth in the presence of NaCl (2%, 4%, and 6.5%), at different temperatures (10°C, 15°C, 40°C, and 45°C), and at pH 9.6. Growth in the presence of bile salt (40%), hemolytic activity (blood agar supplemented with 5% sheep blood), and aesculin hydrolysis (Bile Aesculin Agar, Merck) tests were also performed using isolates from KAA plates.
Gram-positive, catalase-negative, and homofermentative coccus-shaped isolates from M17 Agar plates that could grow at 10°C and 40°C (but not at 45°C or in medium with a 6.5% salt content or pH 9.6) were considered to be possible lactococci. Gram-positive, catalase-negative, and rod-shaped isolates from Rogosa plates (can grow at 15°C and 45°C) were deemed to be homofermentative or heterofermentative lactobacilli. The isolates from MRS+V Agar plates that were Gram-positive, heterofermentative, catalase-negative, and coccus-shaped (arginine hydrolysis negative and no growth at 45°C) were considered to be Leuconostoc spp. Gram-positive, catalase-negative, α- or non-hemolytic and aesculin hydrolysis positive coccus-shaped isolates from KAA plates that could grow in the medium at 10°C–40°C, pH 9.6 and in bile salt were identified as possible enterococci (Navidghasemizad et al., 2009).
To determine the carbohydrate utilization profiles of isolates from M17, Rogosa, Agar and KAA API 50 CH and API 20 Strep systems (BioMěrieux, Inc., France) were used respectively, following the manufacturer’s recommendations. The results were analyzed using APILAB Plus version 1.2.1. (BioMěrieux, Inc., France). Strains were tested in duplicate to determine the reproducibility of the tests. L. lactis ssp. lactis SIK83, Lb. sakei NCDO 2714 and E. faecalis ATCC 29212 were used as control strains.
Genomic DNA was extracted from the isolates as described by Cancilla et al. (1992), and DNA concentration and purity were evaluated spectrophotometrically using a NanoDrop ND-2000 spectrophotometer (Thermo Fischer Scientific, Waltham, MA, USA).
The isolates were genetically characterized by polymerase chain reaction (PCR) of the 16S rRNA gene using the 16S forward (5’-CCG TCA ATT CCT TTG AGT TT-3’) and reverse (3’-AGA GTT TGA TCC TGG CTC AG-5’) primers which are amplifies the 900 bp fragments (Beasley and Saris, 2004). DNA amplification was carried out 50 μL PCR reactions containing 10 μL PCR master mix (Thermo Fischer Scientific), 36 μL nuclease-free water, 1 μL of each primer (100 pmol), and 2 μL (100 ng/μL) of genomic DNA template. Each PCR cycle consisted of an initial denaturation step of 5 min at 94°C followed by 30 cycles of amplification comprising a denaturation step for 30 sec at 94°C, annealing at 55°C for 60 sec, extension at 72°C for 1 min, a single 10 min extension at 72°C for 10 minutes and a final hold at 4°C. PCR products were electrophoresed in 1% agarose gels and then stained with EtBr. An O’GeneRuler 100 bp DNA molecular weight ladder (Fermentas, Finland) was used as a standard. Gels were visualized under UV light using a Kodak Gel Logic 200 Imaging System (Kodak, USA). PCR products were purified using a PCR purification kit (Thermo Fischer Scientific) and were sequenced by MedSanTek (Istanbul, Turkey). Basic local alignment search tool (BLAST) was used to compare the similarity of sequences against the nucleotide database in the National Center for Biotechnology Information (NCBI). L. lactis ssp. lactis SIK83, Lb. sakei NCDO 2714 and E. faecalis ATCC 29212 were used as control strains.
Plate count data were transformed into natural log-based data to perform statistical analyses using SPSS for Windows (IBM, SPSS Inc., Chicago, IL, USA). Significant differences were determined by ANOVA, and mean differences were compared using Tukey’s test at the 5% significance level.
Results and Discussion
The chemical characteristics of the Ezine cheese samples collected from 3 dairies were studied during the manufacturing and ripening periods. Table 1 summarizes the mean values obtained from the physicochemical tests. No significant mean differences in the parameters for the samples (except DM) were detected from different dairies (p>0.05). The observed FDM, pH, and lactic acid in dry matter (LADM) values were consistent with Turkish Cheese Standard 591, which sets the FDM value as at least 45% (w/w), pH≥4.5 and lactic acid content in DM maximum of 3% (TS, 1995). In contrast, SDM values of samples were observed to be higher than that indicated in the standards. The DM contents of the samples collected at different periods were observed to increase significantly (p<0.05) depending on the dairy. Although the DM and FDM values increased from April to June in all samples, a similar pattern was not detected for SDM and LADM. The highest DM and FDM values were detected in the samples taken from B in June. These results are in line with the findings of Oner et al. (2006). Previous studies have shown that the DM and fat contents of cheese are affected by the quality of milk, which is linked to the production conditions and season (Estepar et al., 1999; Oner et al., 2006; Pappa et al., 2016). The mean pH values of cheeses from the three dairies remained constant (p>0.05) during the ripening period. Similarly, the differences in the mean pH values of cheese from different months were also insignificant. Previous studies on starter-free cheeses support these findings (Estepar et al., 1999; Fresno et al., 1996).
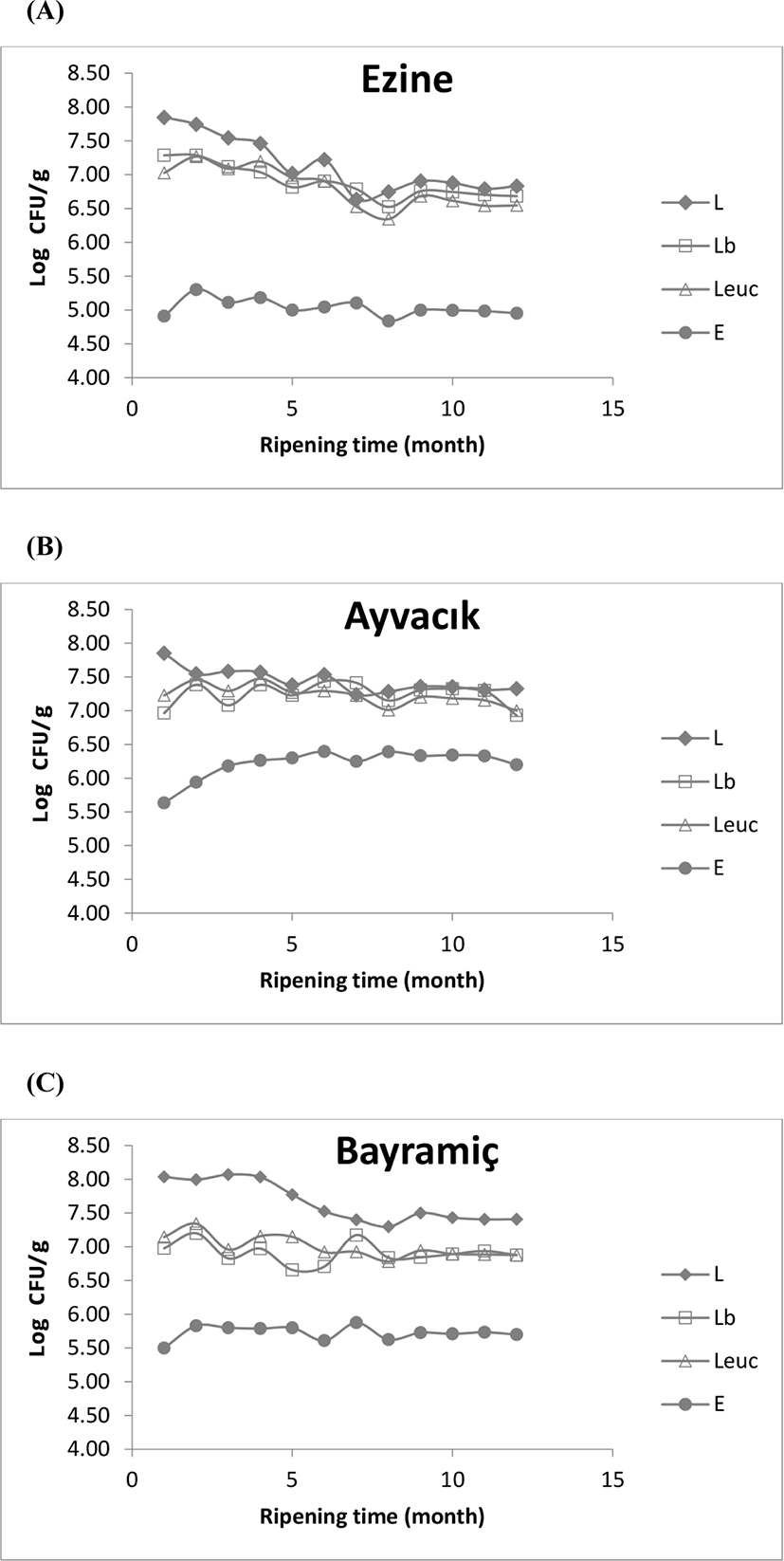
The changes in the microbial groups were analyzed throughout the ripening period. The TMAB counts ranged from 7.01 to 8.16 Log CFU/g and decreased 1-log unit during the ripening process, and the observed YM counts (3.5 Log CFU/g) did not change significantly depending on the dairy and season (p>0.05) (data not shown). Although the TMAB counts observed in this study are in line with the findings of Oner et al. (2006), the YM counts were not. A plausible reason for this difference may be the different ripening periods that were used in these studies. The viable NSLAB counts on different media throughout the ripening period are shown in Fig. 2. The colony counts on M17 plates ranged from 6.00 to 8.18, 6.74 to 7.93 and 7.03 to 8.57 Log CFU/g in dairies E, A, and B, respectively. In detail, the aforementioned counts point out a downward sloping trend in the first nine months of the ripening period, which was greater for E than A and B (p<0.05). In contrast, the colony counts on Rogosa, and MRS+V plates were observed to be similar (ranging between 5 and 7.91 Log CFU/g), and the change in colony counts was not significant (p>0.05). The colony counts on KAA plates ranged from 4.84 and 6.6.4 Log CFU/g, and unlike the other NSLAB groups, increased by 1-log unit during the first nine months of ripening and tended to remain constant afterward. As shown in previous studies, NSLAB is the largest microbial group in all traditionally and industrially produced cheese varieties (Delamare et al., 2012; Domingos-Lopes et al., 2017; Montel et al., 2014; Terzic-Vidojevic et al., 2007). Although the colony counts of the mentioned NSLAB groups above varied, NSLAB remained the dominant flora during the ripening period. Despite the fact the manufacturing environment (i.e., the floors, dairy drains, and equipment surfaces) may be sources of NSLAB, raw milk has been shown to be the primary source of these bacteria (Gobbetti et al., 2015). After the pasteurization process, heat-resistant NSLAB strains remain, recovering and proliferating during the ripening period. Moreover, the ability of some mesophilic LAB to form biofilms is another source of NSLAB, serving as a source of NSLAB through post-pasteurization contamination (Gobbetti et al., 2015; Montel et al., 2014).
One hundred and ninety colonies that exhibited inhibition zones larger than 1 mm against indicator strains were isolated from the various plates. The results of the morphological and physiological characterization assays are shown in Table 2. The isolates from KAA plates grew in the presences of 40% bile salt and on bile-aesculin agar. Also, our results showed that 40 out of 49 isolates from KAA plates exhibited γ-hemolytic activity (data not shown).
According to the phenotypic sugar fermentation profile characterizations, the NSLAB isolates from Ezine cheese belonged to the genera Lactobacillus (36.65%), Enterococcus (25.65%), Lactococcus (23.56%), and Leuconostoc (7.33%). However, Streptococcus spp. (4.71%) and Pediococcus spp. (1.05%) were detected at relatively lower levels, and two isolates could not be identified (1.05%) (Data not shown). NSLAB communities have been described to be primarily composed of mesophilic lactobacilli, such as Lb. paracasei, Lb. plantarum and Lb. curvatus. Nevertheless, enterococci, lactococci, pediococci and micrococci can also be present (Delamare et al., 2012; Montel, et al. 2014; Terzic-Vidojevic et al., 2007).
Several selective media have been used to isolate NSLAB in different studies (Aponte et al., 2008; Estepar et al., 1999; Navidghasemizad et al., 2009). Fox (1999) reported that M17 agar is suitable for isolating lactococci. Although only 11.86% of the strains isolated on M17 agar were assigned to the genus Lactococcus, the remaining isolates were identified as Enterococcus spp., Lactobacillus spp., Streptococcus spp., Leuconostoc spp. and Weissella spp. Most of the strains isolated on Rogosa agar were identified as Lactobacillus spp. (80.0%). This medium is highly selective for mesophilic lactobacilli because of the high acetate concentration and low pH, which suppresses the growth of other LAB. Although the addition of vancomycin to MRS makes the medium highly selective for members of the genus Leuconostoc (Navidghasemizad et al., 2009), Lactobacillus spp., Enterococcus spp., Streptococcus spp., Pediococcus spp., and Weissella spp. were also isolated using this medium. All strains isolated on KAA agar were assigned to the genus Enterococcus. Due to the presence of selective inhibitory components, such as kanamycin sulfate and sodium azide, KAA has been shown to be highly selective for the isolation of Enterococcus spp. In agreement with a previous study by Navidghasemizad et al. (2009), KAA was observed to be the most selective medium for Enterococcus spp. in the present study.
The genomic DNA of 190 isolates was successfully extracted and used to genotypically identify strains based on partial sequencing of the 16S rRNA gene region and was deposited in the NCBI GenBank database. The accession numbers and the level of the antimicrobial activity of all species are provided in Table S1. The isolates were identified at the species level with more than 99% similarity and belonged to seven genera, Enterococcus (60.0%), Lactobacillus (25.26%), Lactococcus (6.84%), Leuconostoc (1.57%), Streptococcus (1.57%), Weissella (1.57%) and Pediococcus (0.52%). In the 12.63% of all isolates were recorded the highest antimicrobial activity (larger than 21 mm), this ratio was found higher in the species of genus Enterococcus (Table S1). Although low pH, temperature, and high salt concentration are stressful conditions for the growth of microorganisms in the cheese environment, NSLAB can grow under these selective conditions. The genetic characterization results of isolates largely reflected that Enterococcus spp. and Lactobacillus spp. were the primary NSLAB groups in Ezine cheese. The 16S rRNA gene sequence analysis results indicated that Enterococcus was the dominant genus. Among the identified isolates, 73, 36, and 5 of them were identified as E. faecium (64.03%), E. faecalis (31.57%), and E.lactis (4.38%), respectively. Forty-eight isolates were assigned to genus Lactobacillus, with 26 isolates identified as Lb. paracasei (54.16%) and 21 identified as Lb. plantarum (43.75%). The isolates assigned to the genera Lactococcus and Leuconostoc were identified as L. lactis ssp. lactis and Leuconostoc mesenteroides, respectively. All Streptococcus spp. isolates were identified as S. lutetiensis, S. infantarius, or S. macedonicus (Table 3).
In this study, inconsistencies between the API and 16S rRNA gene sequence results were observed. The isolates assigned to the genus Lactococcus by API tests were identified as Enterococcus spp. by 16S rRNA gene sequencing. The 16S rRNA sequence results were supported by the physiological test results of the strains isolated from M17 agar, which exhibited growth ratios of 100%, 96.92%, and 74.24% when cultured at 45°C, pH 9.6 or in 6.5% NaCl, respectively (Table 2). According to the 16S rRNA gene sequencing analysis, the most frequently isolated genus was Enterococcus (60.0%), whereas Lactobacillus (36.65%) was observed as the dominant genus of the cheese microbiota according to API tests. The differences between the API and 16S rRNA sequencing data have been described in other studies (Domingos-Lopes et al., 2017; Moraes et al., 2013; Velasco et al., 2004). As stated by Domingos-Lopes et al. (2017), the most plausible justification for these differences might be the loss or gain of plasmids that are essential to the fermentation of sugar, which may lead to metabolic inconsistencies.
LAB are well adapted to the cheese environmental conditions, such as low pH, high NaCl, anaerobiosis, and a lack of fermentable carbohydrates. Their compositions and abundances are primarily related to cheese production technology (raw milk composition, dairy conditions and production period, etc.). However, it is also related to the differences in conditions and length of the ripening period (Montel et al., 2014). Although NSLAB consists primarily of mesophilic facultative and obligate heterofermentative bacteria, lactobacilli, enterococci, pediococci, Leuconostoc spp. and other thermophilic LAB can be members of this population (Gobbetti et al., 2015). Enterococci can be present, grow and contribute the flavors of certain foods, primarily fermented foods, and particularly in cheeses produced in the Mediterranean area that are made from either raw or pasteurized milk from sheep, goats or cows (Fuquay et al., 2011). It has been reported that the count of enterococci in these traditional cheeses may range from 104 to 106 CFU/g in curd, and from 105 to 107 CFU/g in fully-ripened cheese (Giraffa, 2003).
Enterococci are an essential member of the bacterial population of several types of cheese from either ovine or caprine milk such as Cheddar, Feta, Water-buffalo Mozarella, Cebreiro, Venaco, and Hispanico (Giraffa, 2003; Ogier and Serror, 2008; Psoni et al., 2006). Goat and ewe milk is used at a high percentage in the manufacture of Ezine cheese. It was shown that 60% of all species from Ezine cheese were identified as strains belong to the genus Enterococcus. In addition, enterococci have been isolated in high numbers and often represent the predominant microorganisms in brine-ripened cheeses (Navidghasemizad et al., 2009; Fuquay et al., 2011).
According to a report by Fuquay et al. (2011), E. faecalis and E. faecium are the most relevant species in dairy products among the 30 species recognized at present, and E. faecalis typically exhibits higher proteolytic and lipolytic activity than E. faecium or E. durans. In another recent study, Ispirli et al. (2017) reported the presence of Enterococcus species in Turkish white cheese. Additionally, genotypic characterizations of these isolates via 16S rRNA gene sequencing revealed the presence of 12 distinct strains belonging to 5 different species (E. faecium, E. faecalis, E. durans, E. gallinarum, and E. italicus). In this study, 64.03% of enterococcal species were identified as E. faecium, and it was the most commonly isolated enterococci species from Ezine cheese. E. lactis is a recently described novel enterococcal species isolated from Bitto cheese, a PDO raw milk cheese produced in a restricted Italian alpine area (Morandi et al., 2011), as well as from South African fresh sheep milk (Bauer et al., 2009) and Russian sour milk products (Botina and Sukhodolets, 2006). In our study, five species were identified as E. lactis. Due to the importance of the selection of new enterococcal strains other than E. faecalis for the production of traditional cheese in Mediterranean countries, the Ezine cheese originated isolates obtained in this study may be used as novel and safe starter strains. Ambadoyiannis et al. (2005) reported that enterococci isolates from cheese were able to withstand low pH and high bile concentrations while exhibiting antagonistic activities towards other LAB strains. Selected isolates with antimicrobial activity may have both technological and probiotic properties and could be used as adjuncts to produce Ezine cheese. Although IDF has included strains of enterococci in the list of LAB starters (Marino et al., 2003), the virulent nature and antibiotic resistance of most enterococci raise many concerns for the selection of strains as starters.
Lactobacillus is the most abundant genus among LAB, and overall, 18 species of mesophilic lactobacilli have been variously identified (Gobbetti et al., 2015). Among them, the facultative heterofermentative lactobacilli, Lb. paracasei and Lb. plantarum, are dominant in most cheeses. Lactobacillus strains are interesting for the industrial use because of advantages that include faster acidification and resistance to high salt concentrations in long-ripened cheeses (Elortondo et al., 1998; Navidghasemizad et al., 2009). In addition, they have high proteolytic and lipolytic activities, which are essential factors in the development of cheese flavor. It was reported that Lb. paracasei is the dominant species of NSLAB both in several semi-hard cheese varieties (Thage et al., 2005) and in long-ripened cheeses, such as PDO Grana Padano cheese (Levante et al., 2017). In brine-ripened cheeses such as Feta, Teleme, Briza, Halloumi and Domiati, Lb. paracasei ssp. paracasei is one of the predominant lactobacilli (Fuquay et al., 2011). It was reported that Lb. plantarum isolates from Feta cheese, which is a brined ripened cheese similar to Ezine cheese, exhibited lipolytic, and peptidolytic activities (Litopoulou-Tzanetaki and Tzanetakis, 2011). Similar to other studies performed in Italy (Fortina et al., 2003), Northern Europe (Ostlie et al., 2004), the Balkans (Terzic-Vidojevic et al., 2007), Greece (Litopoulou-Tzanetaki and Tzanetakis, 2011) and Portugal (Domingos-Lopes et al., 2017), Lb. paracasei was described as the dominant Lactobacillus flora of Ezine cheese originating from Turkey.
In this study, 6.84% of all strains isolated from Ezine cheese were identified as L. lactis ssp. lactis. Lactococci contribute to the final texture (moisture and softness) and the flavor of dairy products via the activity of their proteolytic and amino acid conversion pathways. Isolates of lactococci differ considerably with respect to their acidifying, caseinolytic, aminopeptidase, and esterase/lipase activities (Litopoulou-Tzanetaki and Tzanetakis, 2011; McSweeney, 2004). L. lactis is naturally dominant in milk and fermented dairy products and is commonly used as starters in the manufacture of most cheese varieties. Rapid lactic acid production of L. lactis in milk has been reported to assist in the formation of curd and flavor, the creation of optimal biochemical conditions for ripening and the prevention of the growth of pathogenic and spoilage bacteria (Fernandez et al., 2011). In agreement with Montel et al. (2014), the primary role of L. lactis ssp. lactis in Ezine cheese may be the decrease in pH, which initiates the coagulation of milk and regulates the early growth of the cheese microbiota during the cheese making process.
Pediococci present only a small proportion of the total LAB in cheeses and are typically not predominant NSLAB (Fuquay et al., 2011). The role of pediococci in cheese ripening is not yet fully understood, although it is known that pediococci work in synergy to affect typical cheese flavors. It is obvious that their role is not as important as that of Lactococcus, Lactobacillus and Enterococcus (Beresford, 2003). In agreement with other studies, in the present study, Pediococcus pentosaceus was isolated at low levels from Ezine cheese. P. pentosaceus isolates from Feta and Teleme cheeses produce acid slowly and often form diacetyl and acetaldehyde and have a proteolytic activity against peptides containing proline (Litopoulou-Tzanetaki and Tzanetakis, 2011). Leuc.mesenteroides was described as the only leuconostoc species isolated from Ezine cheese. It has been suggested that leuconostoc species may enhance the flavor of cheese and contribute to the texture and stabilization of cheese by the production of polysaccharides (Domingos-Lopes et al., 2017). Another non-lactobacilli genus isolated in this study was streptococci. S. macedonicus was first isolated from various artisanal cheeses, such as Greek Kasseri cheese and Italian cheeses, and is known to affect the characteristic flavor of cheese (Gobbetti et al., 2015). In this study, the low level of S. macedonicus observed was in agreement with findings of Lombardi et al. (2004), who sought to observe the technological characteristics of S. macedonicus strains from raw milk cheeses.
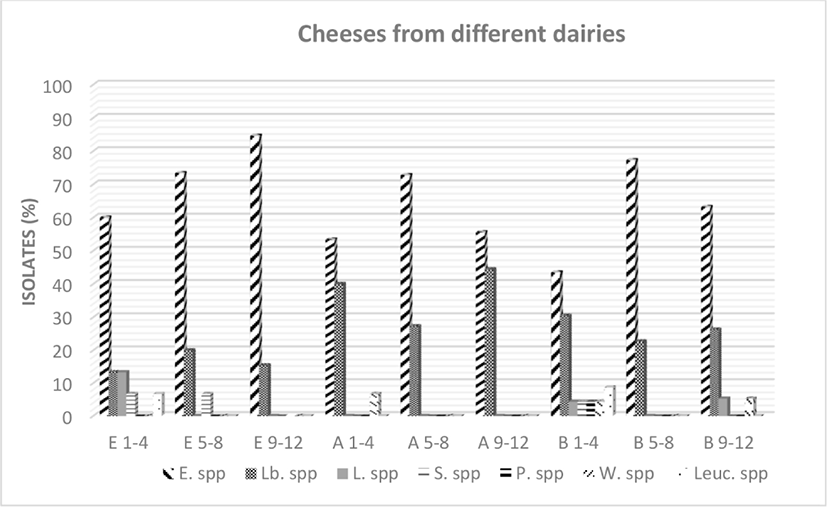
Fig. 3 shows a survey of the NSLAB community in Ezine cheese from each of three dairies during different times in the ripening period. During 12 months of ripening, the dominant flora was observed to be Enterococcus spp. in all dairies. During the first 4 months of ripening, a relatively higher variation in the genus profiles in dairy B was observed compared to the other dairies. The greatest number of Lactobacilli spp. was observed in dairy A and the number of Lactobacilli spp. increased during the ripening period. Unlike enterococci and lactobacilli species, lactococci species were observed to be present in the flora in the first four months of ripening, but not at later periods of ripening. According to these results, the lactation season may affect the NSLAB community in Ezine cheese. As reported by Litopoulou-Tzanetaki and Tzanetakis (2011), the lactation season has a clear effect on the predominance of the certain LAB during each season. Although the primary lactococcal species may be more frequently observed in winter cheeses, facultatively heterofermentative lactobacilli may predominate in spring and summer cheeses. Furthermore, the presence of lactococcal species during the initial months of ripening has been described by Navidghasemizad et al. (2009), Fernandez et al. (2011) and Montel et al. (2014).
The dominance of NSLAB can be attributed to their high tolerance to the hostile environment of cheese during ripening compared to many other microbial groups and to their ability to inhibit undesired microorganisms. This especially results in the microbiological stability of the long-ripened cheeses (Settanni et al., 2010). Ezine cheese appears to be more susceptible to spoilage contaminations since the TMAB and YM counts did not change significantly during the ripening period. The detected high antimicrobial activity in this study indicates that NSLAB may contribute the microbiological stability of Ezine cheese during ripening. During the long ripening periods, the protective features of non-starter flora are important, with microbial spoilage encountered at a minimum level in traditionally produced cheeses. In cheese, the protective nature of LAB against foodborne pathogens and spoilage organisms confer a competitive advantage (Settanni et al., 2010). Therefore, in this study, the isolation of LAB strains with antimicrobial activity from Ezine cheese was the first criterion in the selection of strains with starter culture potential.
Conclusion
To the best of our knowledge, this study is the first report focusing on the characterization of antagonistic NSLAB microbiota from Ezine cheese (PDO), a traditional and widely consumed Turkish white cheese. The results of this study highlight the large biodiversity of the spontaneous LAB microbiota, with 13 different species belonging to seven genera observed. In the present study, the isolation and molecular characterization of NSLAB with antimicrobial activity resulted in the formation of defined collections of natural NSLAB isolates from Ezine cheese. The selected strains that are having technological properties as well as have the ability to produce bacteriocin against pathogenic or spoiling microorganisms could be used for the formulation of specific starter or adjunct cultures for the large-scale production of Ezine cheese (PDO).
In the future studies, the selected isolates will be screened for their ability to grow at non-optimal temperatures; their proteolytic, lipolytic, acidifying and autolytic activities; their antimicrobial activity against Gram-positive and Gram-negative microorganisms; and for the production of acetoin and bacteriocin.