Introduction
Food safety is a major concern for industry and consumers. The pathogenic bacteria are very common in food, and extreme care should be taken to ensure contamination-free meat products for market supply (Newell et al., 2010). Meat is a nutritious and suitable medium for microorganism’s growth, especially the pathogenic one, which subsequently secrete several poisons and cause different food-borne illnesses (Mboto et al., 2012). Incorrect handling and/or storage during meat treatment could reduce the shelf life of meat especially at a certain temperature (Nychas et al., 2008).
Meat color is the most common sensory indicator for the meat quality and freshness. It has been found that the microbial growth could also alter the color of meat-stuffs. The current application of fresh meat color has been extensively studied (Mancini and Hunt, 2005), and enhancing the meat color is critical. The color and microbial load changes are the most critical factors that influenced the shelf life of meat (McMillin, 2008). Due to the fact that meat is a potentially suitable medium for microbial growth (Kang and Kim, 2010), many previous studies have intended to use the some food additives to improve the meat color and microbial safety. Nitrite is one of the most color enhancer extensively used in meat processing. Nitrite can react with Mb to form nitrosylmyoglobin (NO-Mb) which is responsible for the pink color of cured meat. In addition, NO-Mb also has anti-pathogenic and anti-oxidation activities (Sebranek and Bacus, 2007). So, nitrate and/or nitrite salts considered as the main curing agents in the formulation of most fermented sausages and many dry-cured hams, but their carcinogenic effects via reaction with secondary or tertiary amines to form N-nitrosamines in meat proteins (Li and Shao, 2013; Li et al., 2016), limited their industrial applications. For example, adding a surplus of reducing agents, e.g., ascorbate was used to reduce the side effects of nitrate and nitrite (Gotterup et al., 2007). On the other hand, significant efforts have been made to find a suitable nitrite-alternative used in meat-stuffs. However, only a few substitutes have been widely used in the meat products (Armenteros et al., 2012; Djeri and Williams, 2014; Feng et al., 2012). Sodium nitrite cannot be used in any product labelled organic, so natural substitutes are becoming more significant to the meat industrial. There are a few substitutes for sodium nitrite in food such sea salt vinegar, Salt, and sugar are used in cured meat brine for at least 6 d, with no refrigeration needed (Ojakangas, 2005). Recent studies found that some microorganisms could convert the metmyoglobin (M-Mb) to red myoglobin (R-Mb) derivatives, which have potential utilisations to improve the red meats’ color (Leroy et al., 2006; Renerre, 1990). Staphylococcus xylosus (S. xylosus) and Lactobacillus fermentum (L. fermentum) were previously demonstrated to convert M-Mb to red R-Mb derivatives (Li et al., 2016).
Since there is an increasing consumer’s demand for natural products with fewer chemical additives has the potential to be used for improving the red color of meat (Messina et al., 2015). Therefore, finding new and/or novel ingredients as a safe nitrite-replacer is critically needed. Hence, this study mainly aims to evaluate the effect of NO-Hb and LAB on the color parameters, microbial and pathogenic safety of minced beef during refrigerator storage. The instrumental color, the growth of E. coli, Staphylococcus, Pseudomonas, Lactic acid bacteria (LAB), and Salmonella, were therefore evaluated during storage the minced beef.
Materials and Methods
All bacteria including pathogenic and non-pathogenic bacteria such as E. coli (E. coli O157:H7) ATCCBAA-1883, Enterotoxigenic [E. coli (ETEC) ATCC35401], Enterohemorrhagic [E. coli (EHEC) ATCC51657GFP], Staphylococcus aureus (S. aureus) ATCC31888, Staphylococcus argenteus (S. argenteus) JABA32044V6S1, Staphylococcus simiae (S. simiae) CCM7229 (DQ127902), Salmonella Typhi (S. Typhi) ATCC19430, Salmonella Javiana (S. Javiana) ATCC10721, Salmonella Enteritidis [S. Enteritidis] ATCC31194, Streptococcus thermophiles (S. thermophilus) ATCC19258, Lactobacillus bulgaricus (L. bulgaricus) ATCC11842, Lactobacillus casei (L. casei) ATCC393, Montessori enterococcus (M. enterococcus) NFR17393, and Pseudomonas lundensis (P. lundensis) ATCC49968, Pseudomonas mudicolens (P. mudicolens) ATCC4685™, Pseudomonas aeruginosa (P. aeruginosa) ATCC35554 were obtained from National R&D Center for Egg Processing Laboratories, College of Food Science and Technology, Huazhong Agricultural University (HZAU; Wuhan, China). All strains of bacteria were stored at -80°C in Luria-Bertani broth medium containing 15% glycerol. The ratios of bacteria were used the method of (Hutkins, 2008; Schwab and Ganzle, 2011) with the following modifications. The natural contamination rate in meats from 30–390 (Log10 CFU) (Lee et al., 2009; Rouger et al., 2017). LAB such as S. thermophilus, L. bulgaricus, L. casei, and M. enterococcus has five different groups includes: LAB (0:1)=in the absence of LAB and presence of other pathogenic; LAB (1:1)=in the presence of LAB and other pathogenic in the same ratio; LAB (2:1)=in the presence of LAB and other pathogenic, respectively; LAB (3:1)=in the presence of LAB and other pathogenic, respectively; (4:1)=in the presence of LAB and other pathogenic bacteria, respectively.
NO-Hb yield from beef blood was obtained from the College of Food Science and Technology, HZAU. NO-Hb was prepared by a single factor and orthogonal experimental design. The samples were divided into nine equal parts in which each part was randomly assigned to one of the three levels and four factors treatments. They were centrifuged at 8,000, 8,500, and 9,000×g under the homogeneous in aseptic conditions for 15 min. A 200 mL of MillQ-H2O was added to obtain 200 mL of red blood cells under homogenizer mixing speed of 6,000×g for 3 min. The mixture was then filtered with a cloth to remove the collected blood and followed by addition of 600 mL of MillQ-H2O, then vitamin C (VC, 4.0, 5.0, and 6.0 g), and sodium nitrite (NaNO2, 2.5, 3.0, and 3.5 g) were added. The pH of the mixture was maintained at (5.0, 6.0, and 7.0), and later it ventilated with N2 for 30 min to discharge air from the mixture. The UV absorption of NO-Hb prior drying was determined using a NANODROP Spectrophotometer (NanoDrop 2000c, Thermo-Fisher, USA). A 30 mL of NO-Hb solution was diluted to 1,000 mL with MillQ-H2O. A small portion of each sample was placed in a cuvette and then scanned between 450–650 nm. All samples were scanned using the method of (Pittioni, 2001). MillQ-H2O was used as a control.
A 25 kg of frozen beef samples, as raw materials, were collected from Wuhan city slaughters. The samples were aseptically transferred in a sterile polyethylene food bag to our laboratory. The proper amount of frozen beef such as legs and semitendinosus matured muscle parts were thawed for 24 h at 4°C. Visible fats and surface dirt were removed, cut into small chunks, and minced. The meat was minced in special aseptic conditions to avoid cross contaminations. Samples were dived into two parts; the first one was dived into nine equal and contaminated with bacteria as tabulated in Table 1. Each type of part two was divided into four equal parts and randomly assigned to one of cold storage at 4°C for 7, 10, 13, and 16 d after inoculating bacteria with and without NO-Hb. The stored samples were evaluated for their TBC, Staphylococcus, Pseudomonas, E. coli, Salmonella, L. bulgaricus, L. casei, M. enterococcus, and S. thermophilus. Afterward, the minced beef samples were injected with Luria-Bertani broth containing each strain at the ratio of 1 mL/kg of each sample.
TBC, total bacterial count (E. coli, Staphylococcus, Pseudomonas, Salmonella, lactic acid bacteria); E. A, E. coli O157:H7; E.B, E. coli (ETEC); E. C, E. coli (EHEC); St. A, S. aureus; St. B, S. argenteus; St. C, S. simiae; P. A, P. lundensis; P. B, P. mudicolens; P. C, P. aeruginosa; S. A, S. Typhimurium; S. B, S. Javiana; S. C, S. Enteritidis; Un, uncountable=too numerous to count (TNTC): more than 300 colonies; -, not detected; OR, out of range. Log10−1 CFU/g, first dilution; Log10−2 CFU/g, second dilution; Log10−3 CFU/g, third dilution; Log10−4 CFU/g, fourth dilution.
The glassware was washed thoroughly and left to dry (Harrigan, 1998; Lopez-Diaz et al., 2000). The glassware was sterilized in an autoclave at 121°C calculated for at 15 min. Inoculating wires, loops, and metal instruments such as spatulas, knives, and forceps were sterilized by flaming after dipping in ethanol (Hamed, 2017). Each sample (30 g) was homogenized in 270 mL of sterile 0.1% peptone water. The mixture was homogenized using a sterile stomacher bag at 500 rpm (homogenizer Ningbo Scientz Biotechnology Co., Ltd., China). This solution was referred to as a stock solution (dilution 10−1) and decimal dilutions prepared. One mL of the first dilution (10−1) transferred to 9 mL peptone water (10−2) mL, and serial decimal dilution up to 10−6 was prepared as described by Harrigan (1998).
A 15 mL of melted, cooled (45°C-60°C) medium with agar to each Petri dish was added and allowed to solidify. A 0.1 mL of each of the dilutions to the agar surface was transferred. A 0.1 mL portions on the surface of the agar plates utilizing glass spreaders (Drigalsky spatulas) was quickly spread. The surfaces of the plates were dried for 15 min. The plates were incubated in an inverted position, at 37°C for 2–3 d (Yousef and Carlstrom, 2003).
TBC was determined on plate count agar (PCA, Hope Bio-technology Co. Ltd., Qingdao, China) after incubation at 37°C for 48 h. The colonies developing on plates were counted (Haque, 2017; Kim et al., 2016).
A 63 g of Baird Parker agar medium (Hope Bio-technology Co. Ltd., Qingdao, China) was mixed with 950 mL MillQ-H2O. The mixture was boiled to completely dissolve the medium then sterilized in an autoclave at 121°C for 15 min and then cooled to 50°C. A 50 mL of Hi-media egg yolk tellurite emulsion (FD046) were added, well mixed, and poured into Petri-dishes (Jung et al., 2015).
The MacConkey (Hope Bio-technology Co. Ltd., Qingdao, China) broth medium was used to detect the Coliform bacteria. The dilutions 10−1, 10−2, and 10−3 were used. A 1 mL of the diluted samples were transferred into each of the MacConkey tubes (three tubes for each dilution), and one tube is a control. The tubes were then incubated at 37°C for 48 h. The positive results were indicated by the presence of acid and gas in Durham’s tube. A 1 mL from positives pervious tubes was transferred into each of brilliant green bile lactose broth (BGBL) (Oxoid Co. Ltd., Wade Road Basingstoke, Hants, UK) tubes (three tubes for each dilution), and one tube is a control. The tubes were then incubated at 44°C for 24–48 h. The positive tubes were indicated by the presence of acid and gas (Genta and Heluane, 2001; Harrigan, 1998; Spencer and de Spencer, 2001; Yousef and Carlstrom, 2003). The CFB was identified by the method of (Kang et al., 2014). The general procedure detection of E. coli was examined by Rompre et al. (2002). Mixing each test tube had gas was watched at the BGBL identification (ID) method. Then a loopful from BGBL was transferred to the surface of selective medium eosin methylene blue Agar (EMBA) and incubated at 37°C for 24 h to obtain the isolated colonies of E. coli (Kodaka et al., 2006).
The stage of the pre-enrichment of Salmonella was done by mixing 25 g of the samples with 225 mL of buffer peptone water in a sterile microbiological bag. The pre-enrichment culture was incubated for 24 h at 37°C. The stage of the selective enrichment of Salmonella was done by pipetting 1 mL of pre-enrichment culture to l0 mL of selenite cystine broth and incubating at 37°C for 24 h. The stage of the plating on a selective agar medium was done by transferring a loopful of the selective enrichment medium to the surface of the selective bismuth sulphite agar (BSA) (Hope Bio-technology Co. Ltd., Qingdao, China). The BSA plates were incubated at 37°C for 48 h. Typical Salmonella colonies on BSA were appeared brown or grey to black, sometimes with a metallic sheen. The medium surrounding the colony was usually brown at first and then turned black as the incubation time increases.
From suitable dilution of samples, a 0.l mL was aseptically transferred on solidified MRS agar (The Surface-Spread Plate). The sample was spread all over the plate by a sterile bent glass rod. The plates were incubated using anaerobic jars and gas generating kits for anaerobic growth (Cho et al., 2013).
Pseudomonas bacterial was determined on Pseudomonas CFC selective agar medium, incubated using the method of (Hamed, 2017).
The ZE-6000 colorimeter (Nippon Denshoku, Kogyo Co., Tokyo, Japan) was used to measure the color parameters (aperture size of 8 mm, illuminate D65, the incidence angle of 0°). The D65 was used as the standard illuminate and calibrated using a white standard plate (CIE L*=95.26, CIE a*=-0.89, CIE b*=1.18). The minced meat color difference was analyzed using the reflectance mode with a 3-cm port sample-plate. Chroma (C) measures color saturation or intensity, the total color change (ΔE), and the hue angle (H°) describes the relative amounts of redness and yellowness where 0°/360° is defined for red/magenta, 180° for green, 90° for yellow, and 270° for blue or purple color (Ayala-Silva et al., 2005). A lower (H°) value indicates a redder product. (H°) and (C) were calculated from CIE a* and CIE b* values (Korley Kortei et al., 2015), according to the following formula:
The color difference if pre-treated samples, ΔE, was calculated in related to the control sample (Korley Kortei et al., 2015; Saricoban and Yilmaz, 2010) as follows:
Where CIE L0, CIE a*0, and b0 are valuing for initial. CIE L*, CIE a*, and CIE b* values for the final/pre-treated sample.
Browning index (B.I) represents the purity of brown color and is considered as a significant parameter associated with browning (Gao et al., 2014). The BI was calculated using CIE L*, CIE a*, and CIE b* (Korley Kortei et al., 2015; Mohammadi et al., 2008) using the next equations:
NMR relaxation measurements were achieved on a Niumag NMR analyzer (NMI20-015 V-I, Niumag Co., Ltd., Suzhou, China) operating at a resonance frequency for protons of 22.6 MHz. The methods followed those of (Li et al., 2018; Shao et al., 2016a) with minor modifications. Minced beef, approximately 1.04 g sample were first stuffed into a chromatographic bottle of 2 mL volume, then the whole was placed in a 15-mm glass tube and inserted in the NMR probe (Soininen et al., 2009). The spin-spin relaxation time, T2, was measured using the Carr-Purcell Meiboom-Gill sequence (CPMG). Data from 2500 echoes were acquired from 32 scan repetitions. The temperature of the LF-NMR instrument was maintained at 32°C during each measurement which was performed in triplicate. Post-processing of NMR T2 data was distributed by the exponential fitting of CPMG decay curves performed using Multi-Exp Inv Analysis software (Niumag Electric Corporation, Shanghai, China) (Li et al., 2018; Shao et al., 2016a). For a better fit, multi-exponential fitting of relaxation data was then performed with MATLAB (R2011b, Mathworks Inc., Natick, MA, USA) using the Levenberg-Marquardt algorithm (Li et al., 2018; Marquardt, 1963). This analysis resulted in a plot of relaxation amplitude for individual relaxation processes versus relaxation time (Marcone et al., 2013). Three relaxation times (T21, T22, and T23) and their corresponding water population percentages (A21, A22, and A23) were documented (Shao et al., 2016b).
We designed an experiment using three strains of each pathogenic bacteria to further authenticate our results. The experiments were performed in a triplicate. Microbiological analysis was conducted at 0, 7, 10, 13, and 16 d after inoculated bacteria with and without NO-Hb. Data were analyzed and summarized by the main effects for raw, nitrite, NO-Hb, and LAB paired comparisons among them. The data were analyzed using SPSS software (SPSS, Windows version 12.0, SPSS Inc., Chicago, IL, USA). The effects of NO-Hb and LAB indifferent storage periods were tested by two-factor analysis of variance (ANOVA). Significant differences (p<0.05) among means were identified using Least Significant Difference (LSD) procedures.
Results and Discussion
Table 1 shows that the mean Log10 CFU/g value of TBC, E. coli O157:H7, E. coli (EHEC), and L. casei were 0.58, 0.12, 0.02, and 0.25 at 10−1 Log CFU/g, respectively in a fresh beef before injection with bacteria. Staphylococcus, Pseudomonas, Salmonella, L. bulgaricus, M. enterococcus, and S. thermophilus were not detected in a fresh beef before injection with bacteria. In the control samples bacteria in the ratio (1:1) injected in fresh beef (without NO-Hb addition), the TBC, E. coli O157:H7, S. aureus, P. lundensis, S. Typhi, L. bulgaricus and M. enterococcus counts were Uncountable (Un), 2.06, 1.10, 1.61, 1.8, 0.88, and 0.56 at 10−4 Log CFU/g, respectively. After NO-Hb addition, the counts of TBC, E. coli (EHEC), S. aureus, P. mudicolens, S. Typhi, S. Javiana, S. Enteritidis, L. bulgaricus, and M. enterococcus were 2.02, 0.58, 0.56, 0.24, 0.25, 0.52, 0.56, 0.87, and 0.88 at 10−2 Log CFU/g, respectively. From these data, it can be seen that all bacterial counts were significantly decreased after adding NO-Hb (p<0.05). The counts of Salmonella were significantly higher than the other pathogens, suggesting its better resistance against NO-Hb compared to the other pathogens. However, L. casei, M. enterococcus, and S. thermophiles hardly appeared after added NO-Hb. This indicated that LAB such as (L. casei, M. enterococcus, and S. thermophiles) were very sensitive to NO-Hb. These results are well agreed with our recent results (Hammad et al., 2017) and a slight difference with a related study (Liu et al., 2019). NO-Hb formed from sodium nitrite, vitamin C, and beef blood cells have been proven to have the ability to color the cured meat products found the color stability of NO-Hb and its dispersion in solution was ideal. NO-Hb, significantly inhibited the pathogenic bacteria, because it contains sodium nitrite and vitamin C. Our studies showed that in preparation of NO-Hb, LAB addition significantly inhibited the pathogenic bacteria and increased the color stability. The application of NO-Hb and LAB and its solubility in solution was also improved. The results revealed that NO-Hb and LAB produced an acceptable pink color which is similar to the color produced by nitrite and can be used as a colorant to replace nitrite in processed meat products. These results were consistent with a previous study indicating that the addition of NO-Hb to a minced beef would increase meat color (Hammad et al., 2017). (Wang LY et al., 2016) showed that the increase of meat redness due to the conversion of metmyoglobin to bright red myoglobin derivatives in minced beef using LAB.
As shown in Table 2, the TBCs were 2.02 found at 10−4 Log CFU/g before adding NO-Hb. However, TBCs were significantly decreased to reach 0.68 at 10−2 log CFU/g after adding NO-Hb (p<0.05). The combined of E. coli, Staphylococcus, and LAB were 0.34, 2.27, and 2.18 at 10−2 Log CFU/g, respectively, before adding NO-Hb. However, E. coli, Pseudomonas, and Salmonella growth were rarely observed when combined with LAB and adding NO-Hb. These results are agreed well with related studies (Gaggia et al., 2010; Ghanbari et al., 2013). Pseudomonas and LAB counts were about 1.64 and 0.40 at 10−4 Log CFU/g, respectively, after 7 d of cold-storage. The bacterial counts were increased as influenced by storage periods, which was agreed with the results that reported by Gotterup et al. (2007). It was found that the most isolated bacterial counts belonged to the Pseudomonas species. The counts of LAB were of 0.40 at 10−4 Log CFU/g before adding the NO-Hb. However, it was hardly found after adding NO-Hb. The number counts of TBC, Staphylococcus, Pseudomonas, and LAB were 2.79, 0.52, 2.30, and 1.91, at 10−3 Log CFU/g, respectively on 10 d of storage before adding NO-Hb. The TBC and LAB were significantly increased compared with that at 1 wk of storage. The count of E. coli exhibited a positive tendency by extending the storage time. These results disagree with a previous result (Saad et al., 2001). The count of Salmonella was significantly decreased compared with that of storage after 7 d. This results were agreed with related findings (Charlier et al., 2009). There were no significant differences between the count of Pseudomonas after 7 and 10 d of storage. TBC and LAB were significantly decreased after adding NO-Hb (p<0.05). LAB counts were significantly decreased after adding NO-Hb, from uncountable to reach 0.99 at 10−1 Log CFU/g. The combination of LAB itself was more resistance to NO-Hb, compared with other pathogenic. As understood from the finding in this study, it was seen that the intensity of the LAB is lower than that before adding NO-Hb. These results were in agreement with those reported by Li et al. (2016). Furthermore, the use of NO-Hb combined with LAB may be useful to nitrate/nitrite substitute to inhibit the pathogenic organisms.
TBC, total bacterial count, [Escherichia coli, (E. coli O157:H7, E. coli (ETEC), E. coli (EHEC), Staphylococcus, (S. aureus, S. argenteus, S. simiae), Pseudomonas, (P. lundensis, P. mudicolens, P. aeruginosa), Salmonella, (S. Typhimurium, S. Javiana, S. Enteritidis), LAB, (S. thermophilus, L. bulgaricus, L. casei, and M. enterococcus)]; Un, uncountable=too numerous to count (TNTC): more than 300 colonies; -, not detected; OR, out of range. Log10−1 CFU/g, first dilution; Log10−2 CFU/g, second dilution; Log10−3 CFU/g, third dilution; Log10−4 CFU/g, fourth dilution.
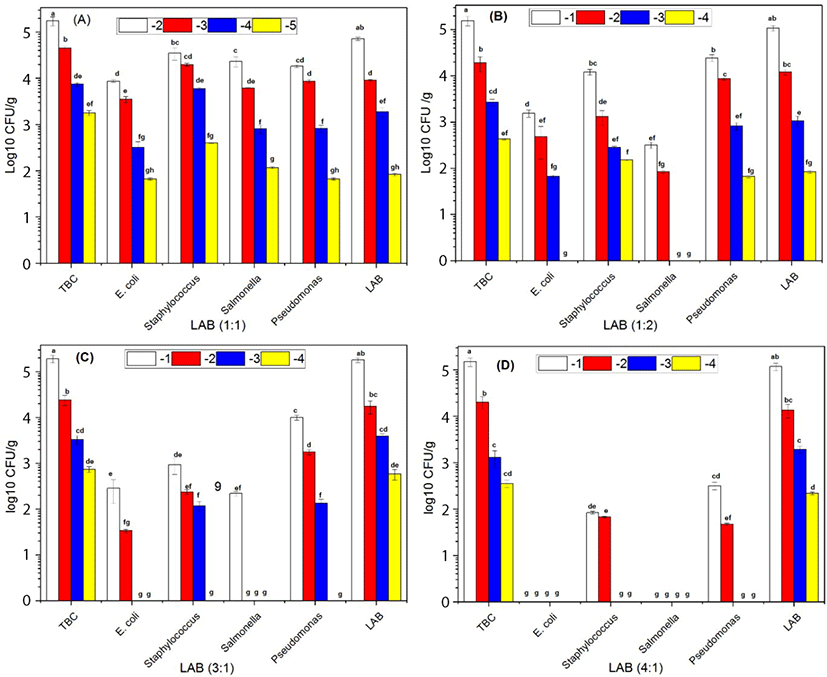
The growth of E. coli, Staphylococcus, and Pseudomonas were not significantly different in the absence of combined of LAB in the ratio (0:1) after 7 d of storage. The growth of the Salmonella (S. Javiana, S. Typhi, and S. Enteritidis) were dominated in the absence of LAB when combined with the others bacteria such as E. coli, Staphylococcus, and Pseudomonas (Fig. 1). The growth of E. coli was decreased compared to Staphylococcus, Salmonella, and Pseudomonas. However, LAB growth became more dormant compared with Staphylococcus, Salmonella, and Pseudomonas in the ratio (1:1) at 7 d storage (Fig. 2A). E. coli and Salmonella counts were decreased compared with Staphylococcus and Pseudomonas. However, LAB growth became more dormant in the ratio (2:1) at 7 d storage (Fig. 2B). E. coli, Staphylococcus, Salmonella, and Pseudomonas counts were significantly different in present combined of LAB. The growth of E. coli, Staphylococcus, and Salmonella were significantly decreased in the presence of LAB in the ratio (3:1) at 7 d storage (Fig. 2C). E. coli and Salmonella exhibited no growth while Pseudomonas colonies appeared more abundantly than Staphylococcus. However, the LAB count in different serial dilutions was significantly (p<0.05) more than overall pathogenic bacteria (Fig. 2D) and increase in the ratio of LAB by 4:1 resulted in complete inhibition of E. coli and Salmonella. Overall, our results suggested that the increased ratio of LAB in the minced beef had an inhibitory effect on the growth of other pathogenic bacteria. Furthermore, the TBC count increased with the increased ratio of LAB. These results were supported by previous findings of (Park et al., 2018; Yang and Moon, 2018) reported that administration of LAB in minced beef inhibiting the growth of bacteria.
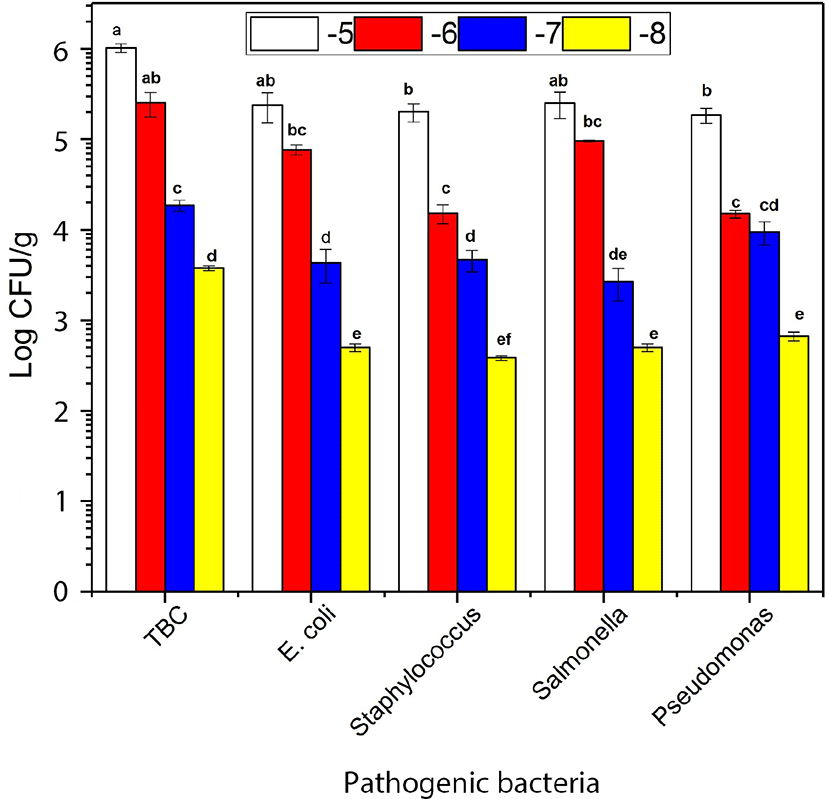
The effect of NO-Hb on the bacterial counts in beef after 13–16 d of storage is shown in Table 3. E. coli, Staphylococcus, and Salmonella counts were of 1.03, 1.27, and 0.43 at 10−1 Log CFU/g, respectively. TBC, Pseudomonas, and LAB counts were 2.52, 1.07, and 1.71 at 10−4 Log CFU/g, respectively after 13 d of storage before adding NO-Hb. The counts of E. coli, Staphylococcus, and Salmonella were significantly decreased (p<0.05) compared with those at 10 d of storage in the presence of LAB. These results were in agreement with the findings of (Affhan et al., 2015). Significant differences in the bacterial counts were observed before and after adding NO-Hb at 13 d of storage. The influence of NO-Hb on pathogenic bacteria in 16 d of storage is shown in Table 3. TBC, E. coli, Staphylococcus, Pseudomonas, Salmonella, and LAB, were 3.73, 0.75, 0.94, 1.57, 0.22, and 2.54 at 10−1 Log CFU/g, respectively, before adding of NO-Hb. The Pseudomonas count was significantly decreased compared to other bacteria. These results were in agreement with those reported by Affhan et al. (2015). These variations of the bacteria growth, especially with the combination of LAB, were statistically significant (p<0.05). These results indicated that the addition of NO-Hb and LAB could inhibit the pathogenic bacteria. And NO-Hb could be used as an unusual ingredient to control the unwanted bacteria in meat and meat-stuffs. Pseudomonas counts were insignificantly increased when the storage period was extended from 7 d to 13 d; however, significantly decreased at 16 d, disagreeing with the results of (Affhan et al., 2015). These results could be attributed to the interference by combining with other bacteria. Moreover, all the bacterial counts were significantly decreased by adding NO-Hb. NO-Hb should be economically or efficiently used in the food industry because the main raw material of NO-Hb is fresh beef blood, which is a kind of abundant resources of iron and proteins of high nutritional and functional quality and then its the price is very low and healthy in comparison of nitrite.
TBC, total bacterial count, Escherichia coli, (E. coli O157:H7, E. coli (ETEC), E. coli (EHEC), Staphylococcus, (S. aureus, S. argenteus, S. simiae), Pseudomonas, (P. lundensis, P. mudicolens, P. aeruginosa), Salmonella, (S. Typhimurium, S. Javiana, S. Enteritidis), LAB, (S. thermophilus, L. bulgaricus, L. casei, and M. enterococcus); Un, uncountable=too numerous to count (TNTC): more than 300 colonies; -, not detected; OR, out of range. Log10−1 CFU/g, first dilution; Log10−2 CFU/g, second dilution; Log10−3 CFU/g, third dilution; Log10−4 CFU/g, fourth dilution.
The total color difference which took the three color parameters lightness (CIE L*), redness (CIE a*), and yellowness (CIE b*) of minced meat samples were determined (Table 4). The results showed that CIE a*- value of minced beef mixed with NO-Hb had insignificant differences (p<0.05) compared with the nitrite samples. The CIE a*- value of the minced beef with NO-Hb was significantly higher than that of either control or the samples inoculated with LAB. The CIE a*-value of the beef inoculated with LAB was slightly higher than that of the control one. Likewise, then continue and significantly (p<0.05) higher than that samples inoculated with LAB combined with other pathogenic bacteria. The CIE a*-value of the beef inoculated with LAB bacteria at different ratios of (0:1), (1:1), (2:1), (3:1), and (4:1) had insignificant differences among them. The visual inspection also showed that the minced beef samples mixed with NO-Hb had a much redder color than that of the samples inoculated with LAB. These results indicated that the minced beef samples mixed with NO-Hb became redder than that of Nitrite, LAB, and other combined bacteria. The CIE L*-values of minced beef samples that mixed with NO-Hb were significantly higher than those of control, LAB, and other combined bacteria. The CIE b*-value showed insignificance among different samples. The redder color in the beef samples mixed with NO-Hb was probably due to the NO-Hb formation. LAB was considered to produce NO-Mb by nitric oxide synthase. The results support the findings of (Zhang et al., 2007). The lower CIE a*-value in the beef with pathogenic bacteria may be attributed to the oxidation of Mb, M-Mb reduction, and acid formation. The results demonstrated the NO-Hb and LAB were capable of producing NO-Mb and inhibition of spoilage bacteria in beef products without the addition nitrite. The results of the color parameters of minced beef and inoculation with bacteria of different ratios during 16 d of storage are presented in Table 4. There was an insignificant difference in luminosity (L-values) of the 0 d, 7 d, and 16 d of storage which ranged from 33.87–44.67, 33.56–45.24, and 33.17–45.24, respectively. However, there were significant differences (p<0.05) among all CIE L*, CIE a*, and CIE b* parameters. In this study, (ΔE) values were calculated concerning the initial time of storage (control meat). The (ΔE) was ranged from 0.80–9.05, 0.2–10.03, and 0.84–8.92 of the 0 d, 7 d, and 16 d storage, respectively. Generally, the lower value of (ΔE) ranging was recorded as the overall color difference, and this could be due to the absence of NO-Hb in meat tissues. The metric (C) was ranged from 21.41–48.7, 21.99–51.23, and 21.63 of the 0 d, 7 d, and 16 d storage, respectively. The (H°) of NO-Hb was 13.01, 13.28, and 13. 35 at 0 d, 7 d, and 16 d storage, respectively. At the 0 d, (H°) reduced to lower values which implied a resultant redder product than the values expressed in 7–16 d, respectively. The (B.I), which was significantly (p<0.05) changed among samples, was in ranges from 21.41–49.78, 21.99–51.23, and 21.63–50.93 at 0 d, 7 d, and 16 d storage, respectively.
CIE L*, lightness; CIE a*, redness; CIE b*, yellowness; (C), Chroma; (H°), hue angle; (ΔE), total color change; (B.I), Browning index; Control, fresh minced beef; Nitrite (100 mg/kg), fresh minced beef with nitrite; NO-Hb (10g/kg), fresh minced beef with nitroso-hemoglobin; LAB, lactic acid bacteria combined such as S. thermophilus, L. bulgaricus, L. casei, and M. enterococcus has five different groups includes; LAB (0:1)=in the absence of LAB and presence of other pathogenic; LAB (1:1)=in the presence of LAB and other pathogenic in the same ratio; LAB (2:1)=in the presence of LAB and other pathogenic, respectively; LAB (3:1)=in the presence of LAB and other pathogenic, respectively; (4:1)=in the presence of LAB and other pathogenic bacteria, respectively.
These results were consistent with those reported by Hammad et al. (2017), Kim et al. (2017), Lei. (2013), and Sawyer et al. (2009), whom reported that the NO-Hb and LAB were increased the meat color.
The effect of different bacteria, refrigerator, and freezing on minced beef is shown in Table 5. Low-field NMR on samples with high water contents generally provides information about the state of the water concerning mobility and distribution of water. From the results, three peaks were identified in the unheated emulsions through the multi-exponential fitting of relaxation data (Li et al., 2018). T21, T22, and A21 represent water, were appeared high in contaminated beef with LAB compared with other bacteria at 0 d. T21 appeared high in contaminated samples with LAB at 7 d storage. A22 appeared high in fresh beef+NO-Hb at frozen storage period for 28 d. The components and population of examined in each water fraction were characterized by the water that exists in macromolecular structures (T21, A21), the water that has more mobility and was trapped within the protein-dense network (T22, A22), the decrease water in the extra-protein network space (T23, A23) refrigerator and frozen storage. The loos water was continued, however, the increased water in the extra-protein network space (A22) in frozen storage at -20°C for 28 d. These results indicated that there was not significantly different between NO-Hb and control in loos water during the storage period. The increasing storage was observed to change both the relaxation times (T2′s) and the distribution (A2′s) of the water in contaminated beef. In all samples, the relaxation time of water that exists in macromolecules (T2 (1, 2, 3)) was observed to be no significantly lower with storage period increased (p<0.05), indicating that the tightly bound water in contaminated beef was intensively trapped. These results were consistent with previous study indicating that the addition of NO-Hb to the minced beef would increase proteins (Shao et al., 2016a).
Values are given as the Mean±SD from triplicate determinations; bacteria in the ratio (1:1) injected in minced beef.
a-f denote refrigerator or frozen in the same column, with different letters differing significantly (p<0.05).
Control, fresh minced beef; Nitrite (100 mg/kg), fresh minced beef with nitrite; NO-Hb (10 g/kg), fresh minced beef with nitroso-hemoglobin; TBC, total bacterial count; Escherichia coli, (E. coli O157:H7, E. coli (ETEC), E. coli (EHEC); Staphylococcus, (S. aureus, S. argenteus, S. simiae); Pseudomonas, (P. lundensis, P. mudicolens, P. aeruginosa); Salmonella, (S. Typhimurium, S. Javiana, S. Enteritidis); LAB, (S. thermophilus, L. bulgaricus, L. casei, and M. enterococcus).
Fig. 3 showed typical examples of the results obtained upon distributed exponential fitting on T2 relaxation data from fresh beef, fresh beef+NO-Hb, beef+nitrite, TBC, and LAB samples. T2 distribution fitted was used to assess the relaxation time of minced beef. Three relaxation components were observed in meat, indicating that the meat proteins restricted the water mobility with different extents. Three components, hereafter referred to as T2b, T21, and T22, were detected in all meat samples: a minor component between 0.1–1.5 ms (T2b), a major component between 10.5–100.25 ms (T21), and finally a third component between 500–750 ms (T22) at zero d of storage (Fig. 3A). T2b and T21 component between (0.8–1.8 ms), and 10.5–150, respectively corresponding to protons that are in macromolecular structures and that are combined closely with macromolecular. However, the main component between T2 (550–750 ms), corresponding to the water or fat that are loosely combined in sol matrix of contaminated minced beef (Shaarani et al., 2006), at frozen storage for 7 d (Fig. 3B). Water holding capacity defines the capacity of minced beef injected bacteria and storage parameter for 14 d. T2b, T21, and T2 component between (1–1.5 ms), (10.3–130 ms), and (450–650 ms), respectively. The relaxation times of the beef protein pellets reduced compared with that at 7 d storage of relaxation times for minced beef (Fig. 3C). The result agreed with (Bertram et al., 2002) who believed that higher water content in the meat might influence the relaxation profiles. Three components, hereafter referred of T2b, T21, and T22, were detected in all beef samples between (0.7–1.7 ms), (15–135 ms), and (135–659 ms), respectively (Fig. 3D). Shorter relaxation time corresponded to a less mobile water fraction, whereas the longer relaxation time corresponded to a more mobile water fraction. The longer the relaxation time represents the less intense the water restriction by the proteins (Bertram et al., 2002; Shaarani et al., 2006). Contrast the beef with bacteria, relaxation time T21 of water group were significantly higher (p<0.05) in meat samples. However, the relaxation time T2b and T2 were not significantly different among treatments. The relaxation components of beef samples were three, indicating that the relaxation components could be caused by water only or a mixture of water and fat. Practically, it was difficult to distinguish between loosely bound fat and strongly restricted water. Consequently, both of them could be accredited to the protons which are tightly combined within macromolecules. Also, the three relaxation time, there were negligible differences among different samples of beef compared with T21 (p>0.05). However, no significant differences of T2b and T2 were observed by the increase of storage period (p<0.05), which indicated that water and fat significantly affected the ability of bacteria. Moreover, the effect of water was undoubtedly higher. The reason could be related to the storage period, bacteria, and storage condition. Similar results have been reported in other studies examining the relaxation time T2 of meat (Bertram et al., 2002).
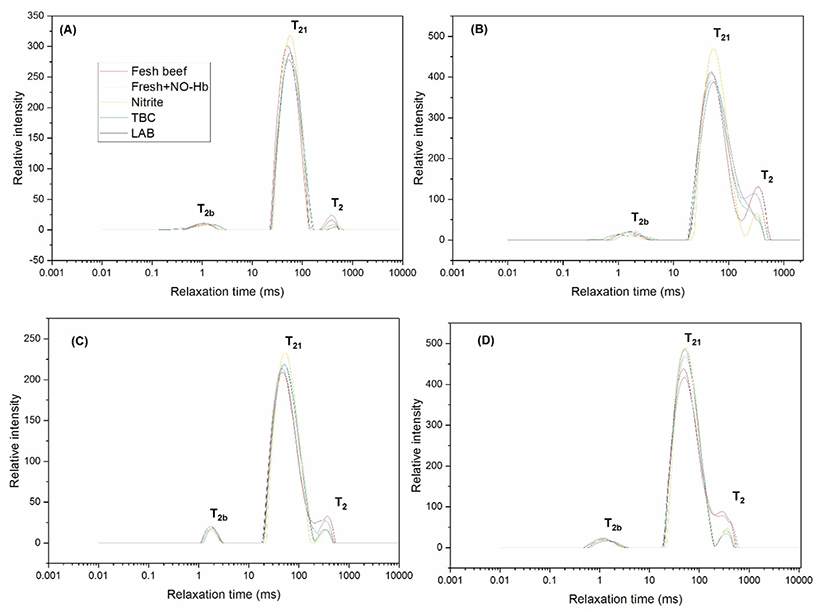
On the other hand, T21 increased after refrigerator storage for 7 d compared at that at 0 d. Also, increase after frozen storage for 28 d compared at that at 14 d, which suggested that the increase of storage influenced the mobility of the water greatly.
Conclusion
This study provides a potential method for nitrite substitution by improving color and inhibiting pathogenic bacteria in beef products. NO-Hb has significant color enhancers and pathogenic inhibitors than that of LAB. Moreover, the use of NO-Hb offer improves the color stability as well as keep the beef relatively more safe than nitrite. The application of NO-Hb as additives in minced beef products is more reliable in preventing foodborne diseases. This study could be extended to improve the shelf life of the meat-stuffs, without affecting the meat color. The use of LF-NMR technique for assessing the influence of water hydration states on the mobility water and fat in contaminated minced beef was investigated. Three relaxation components, T2 (45 ms), T2b and T21 (lower than 20 ms) were found in contaminated beef. The number of relaxation components in minced beef was affected by the increase of storage, but the major component T21 shifted to longer relaxation time (from 500–550 ms) at refrigerator 7 d of the storage. The relaxation time of the same samples was lowered at the frozen stage, indicating that water and fat proton mobility was affected by the type of storage and storage period. LF-NMR has been established to be practicable to identify changes in the relaxation times of water and fat caused by different type of bacteria and the storage periods.













