Introduction
Food hygiene and safety are a major concern in the food industry, and microbiological safety is a particular problem. Escherichia coli can act as an indicator for the presence of other pathogenic bacteria, and it is detected easily in foods such as pork, beef, and chicken. Thus, E. coli detection in foods is one of the most useful hygienic criteria (Scheinberg et al., 2017; Seo et al., 2010; Simancas et al., 2016). However, at present the conventional method for E. coli detection requires several days (Feng et al., 2002; Stromberg et al., 2015; Wang and Salazar, 2016), especially in cases where E. coli concentrations are low. Enrichment is a commonly used method for bacterial isolation to increase the cell counts of target bacteria above other background flora prior to identification (Gracias and McKillip, 2004). According to FDA-BAM (U.S. Food and Drug Administration-bacteriological analytical manual) and other reports, E. coli can be enriched with E. coli (EC) broth or modified tryptic soy broth (mTSB); however, the enrichment methods are time-consuming (Feng et al., 2002; Stromberg et al., 2015).
Polymerase chain reaction (PCR), using primers against the uidA gene that encodes beta-D-glucuronidase can be used to identify E. coli accurately (Molina et al., 2015). PCR detection method has been used to identify a colony on an agar plate, which was formed by plating at least 24-h enriched broth. However, applying PCR detection method directly to the enriched samples has not been evaluated yet. In addition, there is an issue of specificity, since uidA gene is also present in Shigella (Frampton and Restaino, 1993). The objective of the present study was therefore to develop a rapid detection method for E. coli in food samples, using a combination of enrichment and PCR that can also differentiate E. coli from Shigella.
Materials and Methods
Five E. coli strains (E. coli NCCP11142, E. coli NCCP14037, E. coli NCCP14038, E. coli NCCP14039, and E. coli NCCP15661), and Shigella sonnei NCCP14743 strain were cultured in 10 mL tryptic soy broth (TSB, Becton, Dickinson and Company, USA). One-hundred microliter aliquots were transferred to fresh 10 mL TSB, followed by incubation at 37°C for 24 h. The cultures of the five E. coli strains were mixed. Twenty-five milliliters of the E. coli mixture and 10 mL S. sonnei were centrifuged at 1,912 g and 4°C for 15 min, and the pellets were washed twice with the same volume of phosphate-buffered saline (PBS; 0.2 g KH2PO4, 1.5 g Na2HPO4, 8.0 g NaCl, and 0.2 g KCl in 1 L distilled H2O [pH 7.4]). The suspension was diluted with PBS to obtain 3, 4, and 5 Log CFU/mL of inocula, and E. coli and S. sonnei were assayed by PCR to determine the detection limit.
Ham of pork and round of beef were purchased from a butcher shop, and a fresh-cut lettuce was purchased from a supermarket, located in Seoul, South Korea. Ham of pork and round of beef were cut into 20-g portions with a flame-sterilized knife. Fresh pork (20 g, n=4), beef (20 g, n=4), and fresh-cut lettuce (20 g, n=4) were placed aseptically into separate filter bags (3M, St. Paul, MN, USA). E. coli inoculum (0.1 mL) was inoculated onto the surface of the food samples to achieve 1, 2 and 3 Log CFU/g, and samples were massaged 20 times by hand. Samples were then left at room temperature (25°C) for 15 min to allow cell attachment.
Eighty milliliters of EC broth (BD, USA) were placed into the filter bags, and shaken by hand 30 times. All samples were incubated at 44.5°C (Feng et al., 2002) for 0, 4, and 5 h for pork and beef, or 0 and 3, 6, 12 h for fresh-cut lettuce. After enrichment, 1-mL aliquots of the enriched samples were plated onto E. coli/coliform petrifilm (3M, USA) to quantify E. coli. The plates were incubated at 37°C for 24 h, and colonies were manually counted.
One-milliliter aliquots of inocula and enriched samples were centrifuged at 18,341×g at 4°C for 5 min, and supernatants were discarded. Cell pellets were resuspended in 30 µL distilled water and boiled at 100°C for 10 min, and the suspensions were centrifuged at 18,341×g and 4°C for 3 min. The supernatants were then used for PCR analysis.
Primers targeting the uidA and Shigella identification gene were used to differentiate E. coli from Shigella (Table 1). PCR conditions were as follows: 94°C for 2 min (initial denaturation), 94°C for 20 s (denaturation), 72°C for 20 s (extension), and 72°C for 2 min (final extension). Annealing was performed at 53°C for uidA or at 62°C for the Shigella identification gene for 10 s, and 35 cycles were performed. PCR analysis was performed using Fast mix French PCR (i-Taq) (iNtRon Biotechnology, Gyeonggi-do, Korea), and PCR products were run on an agarose gel (1.5%) with electrophoresis for 20 min. Target bands were visualized under UV light.
| Bacteria | Target gene | Size (bp) | Primer sequence (5′-3′) | Reference |
|---|---|---|---|---|
| Escherichia coli | uidA | 252 | PT-2, GCGAAAACTGTGGAATTGGG PT-3, TGATGCTCCATAACTTCCTG |
Cebula et al. (1995) |
| Shigella | Shigella identification gene | 159 | F255, TCGCATTTCTCTCCCCACCACG F413, CCGGATGTGTCTCGGGCAATC |
Kim et al. (2017) |
Results and Discussion
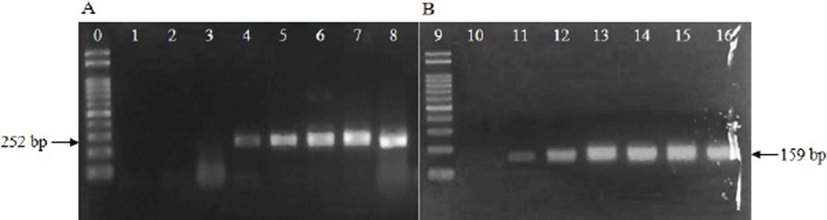
Minimum cell counts for PCR analysis, using primers for uidA gene were 3–4 Log CFU/mL for E. coli and 3 Log CFU/mL for Shigella (Fig. 1). From this result, we confirmed that 3–4 Log CFU/mL of bacterial cell counts was required to detect E. coli with primers targeting uidA gene, and the primers can detect both E. coli and Shigella. Hence, additional primers were necessary to differentiate E. coli from Shigella. Subsequently, the Shigella identification primers described in Table 1 were used, and the Shigella identification primers differentiated E. coli from Shigella (Fig. 1).
Analysis was then performed to determine optimum enrichment times required to obtain 3–4 Log CFU/mL of E. coli for PCR analysis. E. coli was inoculated into fresh pork, beef, or fresh-cut lettuce at 1, 2, and 3 Log CFU/g. E. coli in the pork and beef were enriched for 4 and 5 h, and E. coli in the fresh-cut lettuce were enriched for 3, 6 and 12 h. After 5-h enrichment, E. coli cell counts in the pork and beef increased to 5.9–6.0, 7.1, and 8.0–8.5 Log CFU/g for 1, 2, and 3-Log CFU/g inoculation levels, respectively, and uidA gene expression could be detected at all cell concentrations (Table 2, Fig. 2). In fresh-cut lettuce after 3-h enrichment, the bacterial cell counts increased to 4.2, 5.5, and 6.5 Log CFU/g for 1, 2, and 3-Log CFU/g inoculation levels, respectively, and uidA gene was positive for all samples (Table 2, Fig. 3). Thus, the optimal enrichment time for PCR detection of E. coli was 5 h for fresh pork and beef, and 3 h for fresh-cut lettuce.
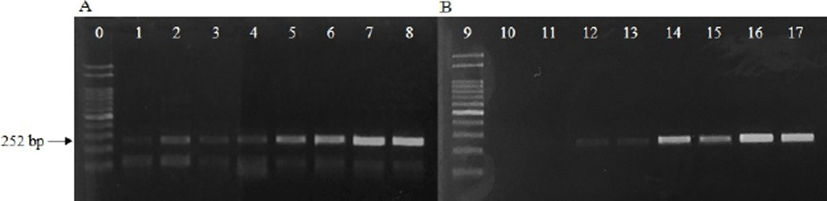
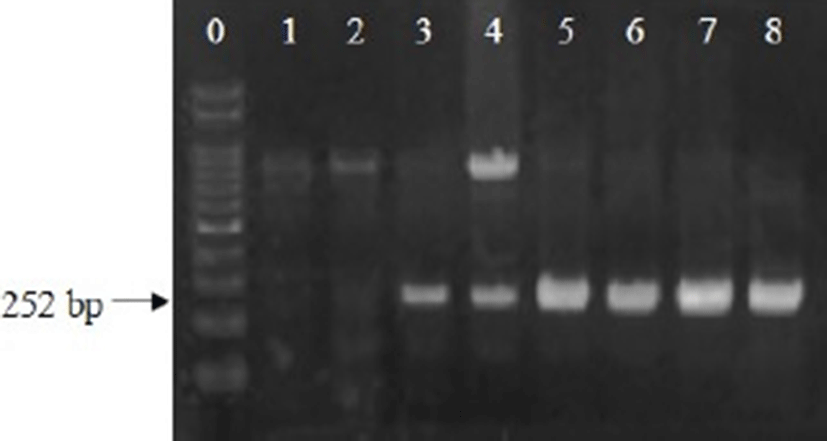
The above results show that meat samples require a longer enrichment time than fresh-cut lettuce. Low E. coli concentrations (0.7–0.8 Log CFU/g) in pork and beef increased to 4.4–4.7 Log CFU/g after 4-h enrichment, and the samples were negative for uidA expression (Table 2). However, at similar E. coli concentrations in fresh-cut lettuce, the samples were uidA positive. It is possible that a component of the meat samples is interfering with the PCR analysis. Wang and Salazar (2016) showed that a number of intrinsic factors can interfere with PCR assays, and other studies have shown that particulates such as fats and carbohydrates can affect nucleic acid amplification (Dwivedi and Jaykus, 2011; Thomas et al., 1991). For this reason, extra pre-treatment, such as centrifugation and bead-based techniques, are necessary to remove some particles from certain foods (Rossen et al., 1992; Yang et al., 2007). Heidenreich et al. (2010) detected E. coli in ground beef using an electrochemical biochip method after enrichment for 4–5 h, and Li et al. (2017) used propidium monoazide treatment to detect viable cell counts of E. coli O157:H7 at 12-h enrichment. However, in this present study, 5-h enrichment for fresh meat samples and 3-h enrichment for fresh-cut lettuce were sufficient to detect E. coli by PCR.
In conclusion, the combination of enrichment and PCR detection method is able to detect E. coli via applying PCR with uidA primers to samples directly after 5-h enrichment for fresh meats (pork and beef) and 3-h enrichment for fresh-cut lettuce.













