Introduction
The issues raising in halal market are interesting in the global economy. It is estimated that the Halal market of food and beverages in the world is expected to increase significantly in the coming years (Owolabi and Olayinka, 2021). It highlights a market size which can be forecasted to increase more than twice from 2023 to 2030, driven by a healthy 10.5% compound annual growth rate (CAGR). From the regional perspective, Asia is the epicenter of halal food market contributing to 59% product share in 2023, since the awareness to consume halal products has increased followed by Europe which holds a 21% product share, while the other regions such as the Middle East, Africa, North- and South America are also key players but with smaller market than Asia (Adekunle and Filson, 2020). According to Maximize Market Research (2025), the commodities of Halal food market can be categorized into some segments, namely meat and poultry, milk and dairy products, fruits, vegetables, cereals, grains, and seafood. Among these, the meat and poultry-based products are dominating Halal market with a market share of approximately 7%. The market of Halal meat globally is estimated to be USD 1,657.44 billion in 2031, increasing at CAGR of 7.4% (Rahman et al., 2024).
The increased halal market of meat products is mainly driven by the fact that Muslims must consume halal foods along with the increased awareness of Muslim to make halal products as their lifestyle. The consumption of halal products as a matter of religious obligation is a must, as stated in the Holy Quran Surah Al Baqarah Verse 168 (Ozturk, 2022). In line with the increased values of global halal market, the market for halal meats and halal meat-based products is also increasing, particularly in Muslim countries like Indonesia, Malaysia and Middle East (Usman et al., 2024). All food products derived from meats are very critical from perspective of halal certification, since meats may come from non-halal animals or they are not slaughtered according syariah principles. Non-halal meat refers to any meat which does not adhere to Islamic law and therefore is not allowed to Muslim consumers. Halal is terms to describe to anythings which are permissible to be consumed or used. In Islamic principle, unless specifically prohibited, every food is considered halal. The opposite of halal is haram (non-halal) which means not allowed and not permissible to Muslim societies (Shahidan et al., 2023).
There are three issues in relation to halal products namely raw materials, processes, and halal authentication analysis. One of the issues encountered in raw materials is meat origins which can be obtained either from its sources (halal and non-halal meats) or due to slaughtering processes (zabiha and non-zabiha). Therefore, some scientist tried to develop analytical methods capable of discriminating the types of halal and non-halal meats due to its sources and its slaughtering process (Muhammad et al., 2019; Nakyinsige et al., 2012). Zabiha slaughtering is the prescribed method of slaughtering animals according to Islamic law to make them halal for consumption. Zabiha slaughtering process must involve the cutting of throat to bring the animal to a quick death without suffering (Addeen et al., 2014).
Halal meats and the products derivatives provide significant sources of nutrition needed for human health, including protein, lipid, vitamins, and minerals, as a consequence, the consumption of halal meats increased recently (Harlina et al., 2022). However, due to its high price, halal meats are often adulterated with non-halal meats for economic purposes. In addition, the main issues related to meats are regarding the procedure of slaughtering process due to ethical, religious, or cultural reasons (Rahman et al., 2024). Some factors that determine whether meat is considered non-halal are:
-
- Meats coming from forbidden animals which are clearly stated in Holy of Quran and hadith of Prophet Muhammad such as pork, canine meat, wild boar meat, and rat meat as well as products that contain or have come into contact with them. Other animals considered haram include birds of prey, carnivorous animals, insects, amphibians, and poisonous fish.
-
- Halal animals such as cattle, chicken, sheep, and goat which are slaughtered not according to Islamic principles. Halal meat must come from the slaughtering process which is according to Islamic law, requiring that the animal to be alive and in good conditions. During slaughtering, the animal must be slaughtered by a Muslim, who uses a sharp knife for cutting the throat of corresponding animal, while reciting the name of Allah. The slaughtering process taken according to Islamic principle is named with “Zabiha”, aiming to minimize the animal’s suffering and ensuring to obtain the meat in good quality (Nurdeng, 2009).
-
- The adulteration practice in which halal meats are substituted or mixed with non-halal meats (Denyingyhot et al., 2022).
-
- The cross-contamination of halal meats from packaging and storage to preparation. Halal products must be clean and free from any non-halal items (Razzak et al., 2015).
The aim of present review was to provide the recent application of several methods to evaluate non-halal meats and meats coming from zabiha and non-zabiha slaughtering processes. During preparing this narrative review, some databases including Web of Science, Scopus, DOAJ and Google Scholar are applied using relevant keywords.
Chemometrics
A basic statistical technique used extensively in many scientific fields, such as halal authentication and food authenticity, is chemometrics or multivariate data analysis (MDA), which analyzes complicated datasets with several interconnected variables. MDA is essential for deriving significant patterns from high-dimensional data in spectroscopic and chromatographic investigations, which improves the identification of minute chemical variations in food items. MDA is an essential tool for trustworthy and data-driven halal authentication procedures since it allows for the simultaneous evaluation of many factors, improving classification accuracy between halal and non-halal components (Nazri et al., 2024; Prihandiwati et al., 2024).
Chemometrics is fundamentally intertwined with MDA, as it involves the application of multivariate statistical and mathematical methods to extract meaningful information from complex chemical datasets (Biancolillo and Marini, 2018). Chemometrics applies statistical and mathematical techniques to chemical data to maximize the extraction of useful information. It is used extensively in analytical chemistry for tasks such as experiment design, instrument calibration, and data analysis. The relationship between MDA and chemometrics is symbiotic, with MDA providing the tools and frameworks necessary for chemometric analysis (Paul and de Boves Harrington, 2021). This integration allows for the comprehensive and accurate interpretation of complex chemical data, driving advancements in various fields such as pharmaceuticals, traditional medicine, and spectroscopy (Qin et al., 2024).
Accurate and trustworthy analytical methods are necessary to guarantee the halal certification of food and pharmaceutical items. Advanced techniques for identifying non-halal components are reported by Nazri et al. (2024) and Prihandiwati et al. (2024), however, their methods, levels of complexity, and areas of application vary. Nazri et al. (2024) stressed Fourier transform infrared spectroscopy-attenuated total reflectance (FTIR-ATR) in conjunction with MDA, whereas Prihandiwati et al. (2024) concentrated on chromatographic and spectroscopic methods for detecting pork products. Based on those studies, the sensitivity and specificity of these approaches are two important differences. Because of the great sensitivity, chromatographic methods like liquid chromatography-mass spectrometry (LC-MS) and gas chromatography-mass spectrometry (GC-MS) may identify traces of non-halal compounds at the molecular level. Because of this, they are perfect for identifying intricate food matrices with extremely low levels of adulterants. These techniques are not practicable for frequent halal verification in industrial settings, though, because they need a lot of sample preparation, costly equipment, and trained workers.
On the other hand, Nazri et al. (2024) showed that FTIR-ATR in conjunction with MDA provides a quick, non-destructive, and economical substitute. Principal component analysis (PCA)’s robustness in classification was confirmed by its ability to discriminate between halal and non-halal samples using the spectral fingerprints of fats and oils, with up to 98.79% variance explanation. This method’s simplicity and low sample preparation requirements make it appropriate for high-throughput screening in industrial and regulatory settings. Nevertheless, spectral databases and chemometric models play a major role in its accuracy, which might result in classification mistakes if the dataset is lacking or if comparable samples have spectral overlap. The capacity of each approach to manage intricate food matrices is another important contrast. Chromatographic methods are more dependable for processed and multi-ingredient items where non-halal adulterants may be chemically changed, since they offer comprehensive molecular information (Nurani et al., 2022). However, because spectral signals are obscured or diluted in highly processed meals, FTIR-ATR may have trouble detecting adulterated bulk fat and oil.
There are functions for both approaches from a regulatory standpoint. While FTIR-ATR is a useful screening method that enables producers to quickly evaluate raw materials prior to more thorough testing, chromatography is essential for confirmatory analysis, guaranteeing halal compliance at trace levels (Irnawati et al., 2023; Nurani et al., 2022). Halal authentication processes might be improved by a hybrid strategy that uses chromatography for confirmatory testing and FTIR-ATR for preliminary screening. Since chemometrics and metabolomics produce massive datasets from high-resolution analytical instruments, the application of MDA has changed dramatically.
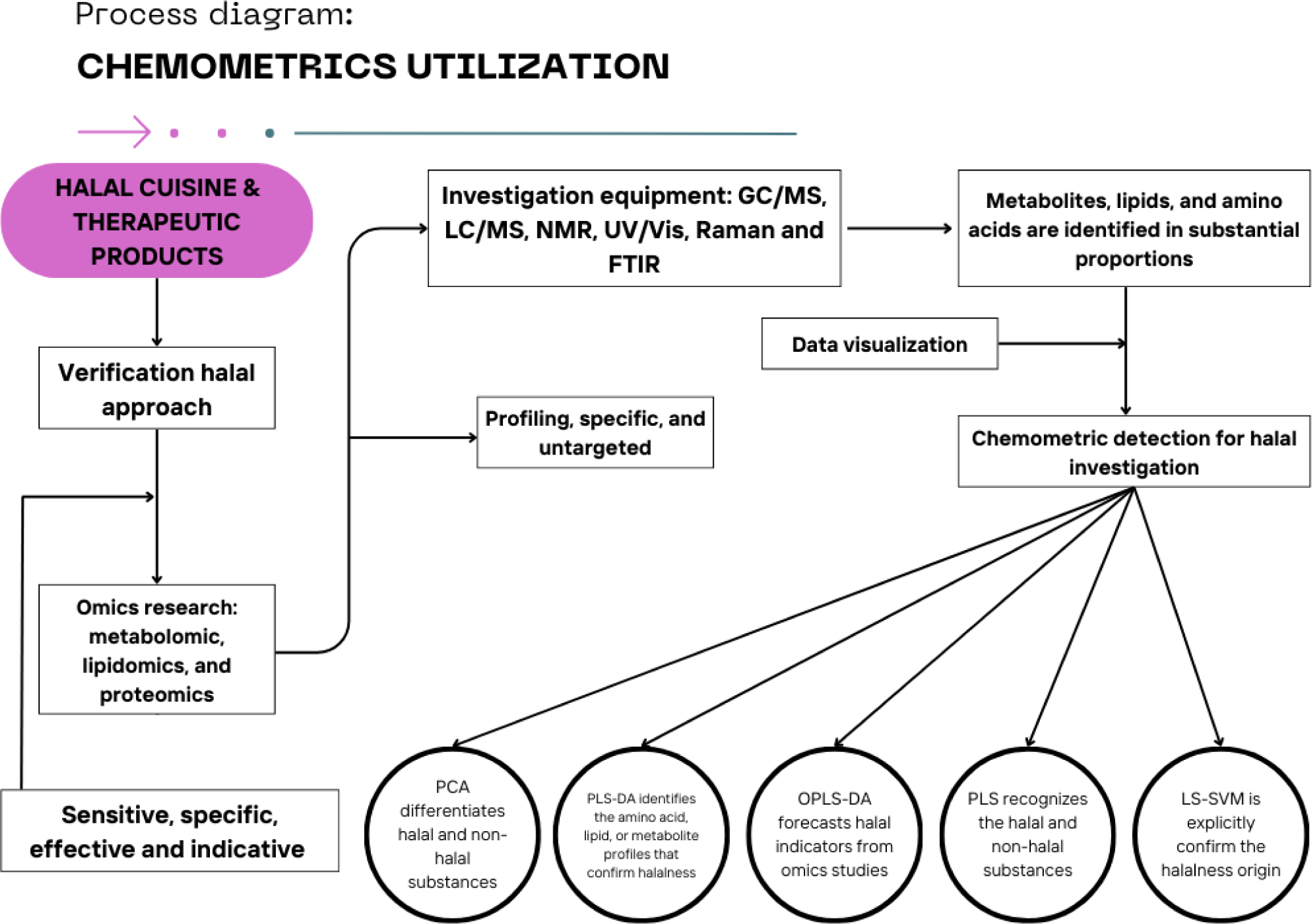
To distinguish between halal and non-halal substances in food and pharmaceutical goods, trustworthy analytical techniques are crucial for halal confirmation (Nurani et al., 2022; Salleh et al., 2022). By identifying distinct biochemical markers, omics-based methods like proteomics, lipidomics, and metabolomics provide great specificity and sensitivity, making it possible to detect minute chemical variations. But handling enormous volumes of data is still difficult and calls for sophisticated computing methods (Musfiroh et al., 2025). Chemometric techniques, such as least-squares support vector machines (LS-SVM), PCA, and partial least squares (PLS), are essential for streamlining complicated datasets, increasing classification precision, and bolstering model dependability. Halal authenticity is further improved by approaches such as partial least squares-discriminant analysis (PLS-DA) and orthogonal projection to latent structures-discriminant analysis (OPLS-DA), which examine differences in metabolic profiles from various sources and slaughtering practices. To increase the precision and effectiveness of halal certification procedures, MDA must be integrated with omics-based methodologies (Dashti et al., 2022; Musfiroh et al., 2025).
According to Musfiroh et al. (2025), omics-based techniques offer accurate molecular insights, which makes them especially helpful for identifying adulteration at the biochemical level. By examining the metabolite profiles, for instance, metabolomics may distinguish between zabiha and non-zabiha meats, while lipidomics can discriminate between fats originating from pork and those that are certified halal. Detecting species-specific proteins in processed foods has also been accomplished with success using proteomics. These methods require specialist knowledge, expensive equipment, and a lot of sample preparation, making them unsuitable for regular halal certification despite their high accuracy. The use of chemometrics in omics research is highlighted in Table 1, and its incorporation into omics-based methodologies is depicted in Fig. 1. However, by identifying significant patterns in big datasets, chemometric methods like PLS-DA and OPLS-DA improve the effectiveness of halal authentication and enable quicker decision-making. By lowering computational complexity, these techniques improve the readability of data visualization and make it possible to forecast possible halal indicators. However, preparation methods and dataset quality have a significant impact on their performance. False positives or incorrect classifications might occur if there are spectral overlaps or if the reference database is lacking. Additionally, for trustworthy verification, chemometrics must be integrated with omics-based techniques as they cannot biochemically Practically speaking, the best method for halal certification is a hybrid strategy that combines chemometrics and omics-based approaches. Chemometric technologies simplify data processing and increase the efficiency of authentication, while omics investigations offer in-depth molecular insights. The accuracy, timeliness, and price of halal certification might be improved by this integrated approach, guaranteeing adherence to Islamic norms while yet being practical for industrial and regulatory uses (Musfiroh et al., 2025). Prior research used univariate methods, which were inadequate for capturing the complexity of meat composition, especially when it came to identifying tainted goods. Strong prediction models that improve the accuracy of meat categorization are made possible by the development of machine learning-assisted chemometrics, which has further improved MDA applications. MDA’s inclusion in halal authentication research ensures accuracy in halal verification techniques and reflects the larger trend in food science toward data-driven decision-making (Maritha et al., 2024; Suratno et al., 2025; Windarsih et al., 2024a; Windarsih et al., 2024b; Yuswan et al., 2019).
| No | Issues | Chemometrics utilization in omics exploration | Outcomes | Ref. |
|---|---|---|---|---|
| 1 | Discrimination of halal (chicken, beef) and non-halal meats (wild boar meat) | Principal component analysis (PCA), partial least squares-discriminant analysis (PLS-DA), orthogonal projection to latent structures-discriminant analysis (OPLS-DA), and heatmap analysis | 1. Metabolites were effectively detected in beef meat (BM), canine meat (CM), and wild boar meat (WBM) by liquid chromatography-high-resolution mass spectrometry (LC-HRMS). 2. The three varieties of meat were successfully distinguished using chemometrics. 3. Important differentiating metabolites for BM-WBM and CM-WBM were found. 4. A potential technique for meat verification that guarantees quality, safety, and halal certification is the combination of LC-HRMS and chemometrics. |
Windarsih et al. (2024b) |
| 2 | Authentication of chicken meat from different slaughtering procedures | PCA, cluster analysis, correlation network analysis, and PLS-DA | 1. Using chemometrics and ultra high-performance liquid chromatography-high-resolution mass spectrometry (UHPLC-HRMS), chicken flesh was effectively verified using slaughtering techniques. 2. A total of 28 distinct metabolite profiles were found, with 3-methylhistidine, creatine, and carnosine serving as the main differentiators. 3. Halal, non-halal, and shubha-halal chicken flesh were successfully categorized and distinguished using chemometric approaches. 4. Thirteen possible biomarkers that can be utilized for trustworthy halal verification were found using PLS-DA. 5. In order to safeguard consumers against non-halal and shubha-halal items, the study validates untargeted metabolomics as a precise and selective technique for confirming the halal quality of chicken flesh. |
Maritha et al. (2024) |
| 3 | Authentication of porcine, bovine, and goat bone gelatins | PCA, cluster analysis, and PLS-DA | 1. By effectively differentiating gelatin origins using UHPLC-HRMS and chemometrics, gelatin’s halal certification was guaranteed. 2. Gelatin generated from goats, cows, and pigs showed distinct metabolic variations, as demonstrated by PCA and cluster analysis. 3. By identifying 15 important metabolites, PLS-DA made it possible to accurately authenticate the provenance of gelatin. 4. The study validated metabolomics as a very successful and selective method for confirming gelatin’s halal certification in culinary and cosmetic applications. |
Harlina et al. (2024) |
| 4 | Chemometrics-identification of potential peptide markers of pork, beef and chicken | PCA and OPLS-DA | 1. Species-specific peptide indicators for identifying pork adulteration in halal foods were effectively discovered using chemometrics-assisted shotgun proteomics. 2. Reliable classification was ensured using PCA and OPLS-DA, which successfully differentiated peptide profiles between chicken, beef, and pork. 3. The fundamental structure of the selected peptide markers was confirmed by the consistent and complementary findings obtained from the combination of peptide mass fingerprinting (PMF) and targeted tandem liquid chromatography-mass spectrometry (LC-MS). 4. This work provided a scientifically proven method for identifying pork contamination in beef and poultry, establishing a reliable and repeatable strategy for halal meat certification. |
Yuswan et al. (2018) |
| 5 | Halal authentication of triceps brachii (TB), longissimus dorsi (LD), and biceps femoris (BF) of meat muscles | PCA, cluster analysis, and PLS-DA | 1. Different metabolic patterns in the TB, LD, and BF muscle regions of beef and pork were effectively discovered by untargeted metabolomics. 2. Meat samples were correctly categorized by chemometric analysis according to their metabolite makeup. 3. Important metabolite indicators that accurately identify halal meat were found using PLS-DA. 4. The work strengthens halal certification techniques by offering a data-driven strategy for differentiating between beef and pork. |
Maritha et al. (2023b) |
| 6 | Analysis of dog meat (DM) adulteration in beef meatballs (BM) using non-targeted | PLS-DA, PLS, and orthogonal PLS | 1. Halal BM were successfully validated using chemometrics and non-targeted LC-HRMS metabolomics. 2. The technique ensured great sensitivity by detecting DM adulteration down to 0.1% (w/w). 3. Biomarker metabolites that differentiated samples containing DM from BM were found. 4. Pathway analysis showed that DM adulteration considerably changed the metabolism of ether lipids, histidine, and beta-alanine. 5. The method’s practical usefulness was demonstrated by its effective application to commercial meatball samples (n=28). 6. This work supports halal authentication efforts by highlighting a practical and dependable method for identifying non-halal adulteration in halal beef products. |
Windarsih et al. (2024a) |
| 7 | Analysis of pork in beef sausages | PCA, PLS-DA, PLS, orthogonal PLS, and variable importance for projection analysis | 1. The metabolite variations between real and fake BS were successfully identified by LC-HRMS untargeted metabolomics. 2. Reliable classification was ensured by the precise separation of BS from pork-adulterated samples by PCA and PLS-DA. 1. PLS and OPLS showed excellent accuracy and low error in their predictions of the degree of pork adulteration with r2>0.99 and root mean square error of calibrations (RMSEC) 1.32%. 2. Eight important metabolites were shown to be biomarkers for the identification of pork, including oleamide, α-eleostearic acid, and arachidonic acid. 3. A reliable technique for identifying pork adulteration in beef sausages was LC-HRMS in conjunction with chemometrics. |
Windarsih et al. (2023) |
| 8 | Detection of pork in tuna meat for halal authentication | PCA and PLS-DA | 1. Pork-adulterated TM was successfully separated from genuine samples using LC-HRMS untargeted metabolomics in conjunction with PCA and PLS-DA. 2. 21 metabolites were shown to be promising indicators for the identification of pork. 3. Pork-specific peptide markers, such as FFESFGDLSNADAVMGNPK (beta-hemoglobin), HDPSLLPWTASYDPGSAK (carbonic anhydrase 3), and HPGDFGADAQGAMSK (myoglobin), were identified by proteomics analysis. 4. Pork adulteration was effectively identified by both proteomics and metabolomics with a sensitivity of 0.5%. 5. This work suggests using LC-HRMS-based proteomics and metabolomics as a standard operating procedure for identifying non-halal adulteration in meat products and validates it as an efficient and trustworthy technique for halal authentication. |
Suratno et al. (2023) |
Many academics concur that very sensitive and selective analytical methods that can identify non-halal adulterants are necessary to guarantee the halalness of meat products. To enhance halal verification, they have used a variety of techniques, such as chemometrics, proteomics, and metabolomics. Although each technique has proven to be successful, there are differences in their precision, applicability, and compatibility for various food matrices, therefore a careful comparison is required to identify the most effective strategy for regular halal certification. Suratno et al. (2025) and Yuswan et al. (2019) employ proteomics-based techniques that concentrate on identifying protein and peptide markers specific to non-halal beef sources. Suratno et al. (2025) modified trypsin digestion settings for a quicker and easier proteomic workflow, whereas Yuswan et al. (2019) improved Gel-Enhanced LC-MS with PCA to reduce human error in protein selection. Because proteins are species-specific and processing-resistant, these methods provide excellent specificity. Proteomics is less useful for identifying adulteration in processed halal meats like sausages or meatballs, nevertheless, because its sensitivity may decrease in highly processed meat products where proteins break down. However, metabolomics-based methods, like those used by Maritha et al. (2024) and Windarsih et al. (2024b), use chemometrics in conjunction with liquid chromatography-high-resolution mass spectrometry (LC-HRMS) to pinpoint important metabolic distinctions between halal and non-halal meats. These investigations proved the method’s better detection threshold by effectively detecting adulterants at a sensitivity of 0.1% (w/w). Furthermore, metabolomics is helpful in differentiating between halal, non-halal, and shubha-halal (something that is potentially halal but contains elements of doubt or uncertainty regarding its permissibility) meats since it may detect molecular alterations brought on by slaughtering practices (Maritha et al., 2024). However, the need for a well-established spectrum database and substantial data processing may restrict metabolomics applicability for everyday commercial application.
All investigations used several chemometric models, such as PLS-DA, PCA, and OPLS, which were essential data analytic tools for distinguishing between real and tainted meat samples. Yuswan et al. (2019) employed PCA to improve protein marker selection, whereas Maritha et al. (2024) and Windarsih et al. (2024b) successfully classified meat samples based on metabolite profiles using chemometrics. Chemometrics’ strength is its capacity to manage intricate datasets while enhancing classification precision. Nevertheless, because it depends on high-quality spectral data, any overlapping spectral signals or absent reference markers may result in misclassification, which would lower dependability in practical applications. We may conclude from the aforementioned research that each approach has particular uses based on the food matrix and halal authenticity specifications. Because species-specific proteins are preserved in fresh meat, proteomics is perfect for authenticating it. However, it might not work well for highly processed foods. Metabolomics provides a highly sensitive and flexible method that can distinguish between different slaughtering techniques and detect adulterated raw and processed meat, making it a more complete solution. In contrast, chemometrics improves both methods but is heavily reliant on reliable spectral databases and necessitates computational know-how for data interpretation.
PCA, one of the most used MDA approaches, is perfect for exploratory data analysis since it preserves important variation while reducing the dimensionality of huge datasets. Based on their distinct metabolite profiles, it has been effectively used to differentiate halal-certified meats from non-halal meat sources such as wild boar, pork, and others (Amalia et al., 2022; Hamdan et al., 2024). By optimizing variance between sample groups, supervised methods such as PLS-DA and OPLS-DA provide improved classification accuracy. These methods are especially helpful in halal authentication studies where minute chemical differences need to be found (Irnawati et al., 2023; Windarsih et al., 2024b). In summary, MDA plays an essential role in identifying non-halal meats since it improves the precision and dependability of analytical findings. Meat adulteration, whether deliberate or accidental, may be accurately identified by its combination with metabolomics and chemometrics. It is anticipated that future developments in MDA, especially the use of machine learning, would enhance the effectiveness and resilience of halal authentication techniques.
Identification and Confirmation of Non-Halal Meats
Numerous analytical techniques have been developed for the identification and confirmation of the meat types (halal or non-halal) and differentiation of meats from zabiha (according to syariah principles) and non-zabiha (not according to syariah principles) slaughtering processes. The most common techniques used are DNA-based methods (Orbayinah et al., 2019) and chromatographic-based methods, as outlined in Table 2 (Windarsih et al., 2022).
| Technique | Description | Advantages | Disadvantages | Ref. |
|---|---|---|---|---|
| LC-MS/MS | LC-MS/MS is used for metabolomics analysis to identify non-halal meats by detecting specific metabolites and protein markers. | 1. High sensitivity and specificity 2. Can detect multiple species markers simultaneously |
1. Requires sophisticated data analysis tools like chemometrics 2. High cost and complexity |
Dewi et al. (2023) |
| Two-dimensional GC-MS | GC-MS is used to detect fatty acid methyl esters and volatile compounds in meat products to identify non-halal meats | 1. High accuracy in identifying specific chemical compositions 2. Effective for processed foods |
1. Time-consuming and requires extensive sample preparation 2. High capital cost |
Iqbal et al. (2025) |
| NMR spectroscopy | NMR spectroscopy is used to profile and fingerprint chemical markers, such as lipids, to identify non-halal meats. | 1. Non-destructive and provides detailed molecular information 2. Can confirm chemical structures |
1. Lower sensitivity compared to LC-MS/MS and GC-MS 2. Requires large sample sizes |
Fadzillah et al. (2017) |
LC-HRMS provides high reliable results in terms of good sensitivity, precision, and selectivity for the analysis of various types of analytes (Lucci et al., 2017). The instrument is divided into ESI+ and ESI– making it capable of separating various types of polar, semi-polar, and non-polar compounds. This instrument is capable of separating and detecting compounds with high accuracy. The HRMS detector has the ability to detect compounds with small molecular weight so that the use of this instrument is very broad, including in omics analysis approaches (Pezzatti et al., 2020). The analysis of metabolites, lipids and peptides can be analyzed accurately to provide a complete profile of the sample. Metabolites, lipids, and peptides that have a small molecular weight with a large number can be analyzed properly using the database in this instrument. This instrument can be applied to the analysis of halal and non-halal meat (Al Olan and Yossouf, 2023).
Analysis of various types of meat including non-halal meat such as pork, rat, and dog can be done by LC-HRMS (Fathima et al., 2024). Metabolites, lipids, and peptides in each species have different characteristics, so they can be used to analyze various types of meat. The results of LC-HRMS analysis of metabolite compounds such as carnitine, DL-malic acid, and leucine were well detected (Maritha et al., 2023b). Analysis of pork, which is similar to beef, was able to be separated by LC-HRMS based on the type of lipids. The lipids present in pork are different from the lipids present in meat in ESI+ and ESI–. This result shows that this instrument can accurately analyze halal and non-halal meat types (Maritha et al., 2023a). Not only non-halal meat can be detected by LC-HRMS but processed meat products and non-halal meat derivatives such as gelatin can also be analyzed using this instrument.
Non-halal processed meat products such as meatballs and corned beef are more difficult to analyze compared to raw meats (Hossain et al., 2023). This is because these products have undergone various processing such as mixing with other ingredients and heating which can cause their composition to change. Non-halal processed meat products will still be able to be analyzed using LC-HRMS through a metabolomics approach, where the metabolites produced will appear to have changed (Hamsar et al., 2024). The high sensitivity of LC-HRMS will be able to detect changes in metabolites present in processed non-halal meat products (Pirhadi et al., 2024).
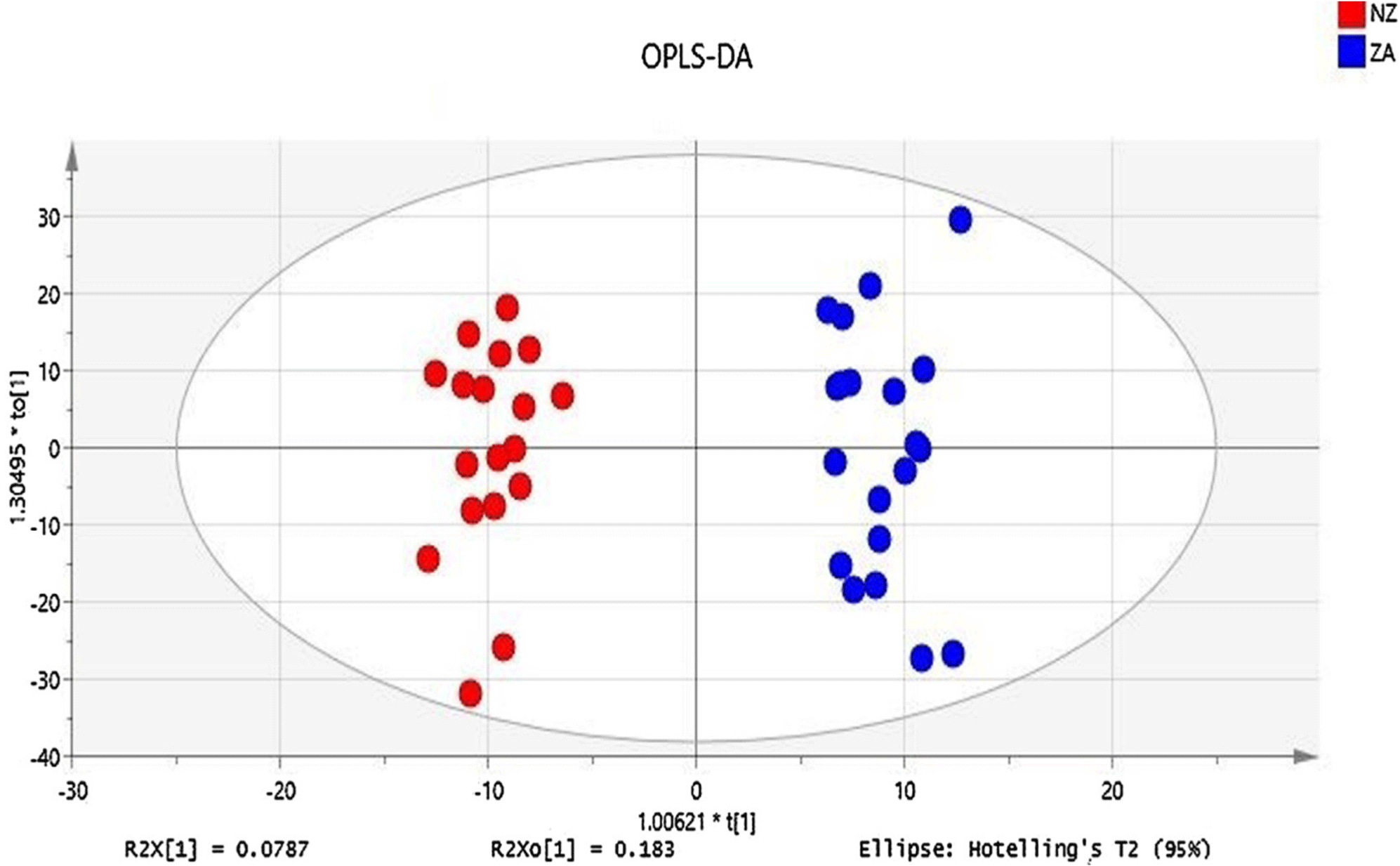
In Indonesia and other countries, chicken meat is the most common meats used as protein sources. The different slaughtering process can affect the changes in chemical compositions of meats, including metabolite profiles (Aini et al., 2023), therefore analytical methods capable of detecting all metabolites have been reported for differentiation of meats slaughtering with zabiha and non-zabiha procedures. Liquid chromatography-electrospray ionization-tandem mass spectrometry (LC-ESI-MS/MS) combined with chemometrics of pattern recognition based untargeted metabolomics has been used for differentiation of 40 chicken meat samples based on slaughtering procedures. The chicken meat samples were classified into zabiha or halal slaughtering (cutting neck without detaching spinal cord) and non-zabiha or non-halal slaughtering. A volcano plot reveals at least 150 metabolites were identified, in which 5 identified and 25 unidentified metabolites could be markers for the clear differences (Abbas et al., 2020). PCA plot on all extracted features does not provide the meaningful separation zabiha and non-zabiha, while OPLS-DA using peak intensities of these metabolites (five identified and 25 unidentified metabolites) could accurately classify both samples with excellent performance (r2 of 0.966, Q2 of 0.609, CV ANOVA p<0.0013), as shown in Fig. 2.
Aini et al. (2022) have used the combination sodium dodecyl sulfate polyacrylamide gel electrophoresis and HRMS equipped with Proteome Discoverer software to evaluate the differences of rat meat proteins slaughtered with zabiha dan non-zabiha procedures. Some changes in protein expression were observed during non-zabiha slaughtering process in which 13 proteins were up-regulated and 3 proteins were detected specifically, namely NSFL1 cofactor p47, transketolase, and Von Willebrand. In addition, there were 3 stable peptides which can be used as peptide markers in those three proteins namely SYQDPSNAQFLESIR related to NSFL1 cofactor p47, LGQSDPAPLQHQVDVYQK related to transketolase, and VPLLCTNGSVVHHEVINAMQCR corresponding to Von Willebrand. Furthermore, Ali et al. (2020) have successfully used ultra high-performance liquid chromatography-time of flight-mass spectrometry (UHPLC-TOF-MS) to identify the different metabolomes in broiler chicken slaughtered in zabiha and non-zabiha manners. Some metabolomes namely glucose, amino acid, inosine, hypoxanthine and arginine were found in different concentrations. These results indicated that LC-MS/MS could identify protein and peptide markers used for identification of meats subjected to the different slaughtering process.
In recent study, Maritha et al. (2024) have used ultra high-performance liquid chromatography-high-resolution mass spectrometry (UHPLC-HRMS) coupled with chemometrics as reliable method for verifying the slaughtering process of chicken meat as halal (zabiha), shubha and non-halal (non-zabiha). Twenty-eight metabolites have been identified in chicken meats for differentiation of chickens slaughtered according to syari’ah principle (zabiha) and non-zabiha. PCA and cluster analysis could effectively cluster chicken meat based on similarities in metabolite profiles. Supervised pattern recognition of PLS-DA could accurately identify thirteen potential biomarkers for halal authentication study, such as creatine, betaine, and acetyl-L-carnitine (Maritha et al., 2024).
Two-dimensional gas chromatography-gas chromatography-mass spectrometry (GC×GC-MS) is an advanced analytical technique gaining prominence in the halal analysis of meat products. This method offers enhanced separation and sensitivity for the detection of non-halal components in food products (Magagna et al., 2016). In the context of halal certification, GC×GC-MS can effectively identify non-halal components, ensuring that products meet the stringent requirements of halal standards (Nurani et al., 2022; Rohman and Windarsih, 2020). The ability to separate complex mixtures with high resolution and analyze volatile compounds provides a powerful tool for confirming the halalness of meat products. Additionally, its high specificity minimizes the risk of false positives, offering a reliable means for regulatory agencies, producers, and consumers to confirm the halal status of meat products (Kaldeli et al., 2024).
The volatile compounds (aroma) in non-smoked bacon were identified using GC×GC-TOF-MS. Some volatile compounds like hexanal and 3-methyl-butanoic acid were identified as key aroma compounds in non-smoked bacon. Moreover, hexanal and 1-octen-3-ol exhibited relatively high flavor dilution factors and odor activity values, therefore, they are considered as the primary contributors. Among the non-smoked bacons obtained from different locations, seven volatile compounds were detected to be contributed to the unique aroma. Therefore, a comprehensive volatile compounds analysis in non-smoked bacons could reveal the potential biomarkers of non-smoked bacons (Wu et al., 2024). In addition, two dimensional GC coupled with TOF-MS also has been successfully used to analyze metabolites, especially fatty acids of pork in various types of food samples (Xu et al., 2015).
The GC×GC-TOF-MS was successfully used to identify volatile organic compounds in pork obtained from Guangdong. The GC×GC-MS could be used to profile different volatile compounds in pork. The observed flavors mainly from aldehydes, ketones, alkanes, and ethers groups. The main observed compounds were octane, pentane, methyl isobutyl ketone, and 3-ethyl-2, 2-dimethylheptane. Therefore, this study revealed the unique flavors of pork obtained from Guangdong which can be used as the identity to detect and recognize pork in halal meat and meat products (Wang et al., 2022).
Nuclear magnetic resonance (NMR) spectroscopy is a powerful analytical technique for detecting non-halal meat and identifying adulteration in meat products. NMR is non-destructive in nature, allowing for the direct analysis of complex mixtures without the need for sample preparation, making it an excellent choice for halal verification (Fadzillah et al., 2017; Gholkar et al., 2021). It can provide detailed information about the molecular composition of meat products, including the presence of non-halal animal derivatives such as pork, which have distinct molecular profiles. By analyzing the chemical shifts and signal intensities in NMR spectra, it is possible to identify specific biomarkers or metabolites that are unique to non-halal species. This ability to differentiate between various animal types based on their molecular signatures makes NMR a valuable tool for verifying the halal status of meat products (Jeong et al., 2020). Furthermore, NMR can detect both the presence of specific non-halal substances and the potential contamination of meat products, ensuring food integrity and compliance with halal certification standards (Akhtar et al., 2021).
Analysis of pork adulteration in meat and meat products was successfully performed using NMR spectroscopy in combination with chemometrics of PCA, OPLS-DA, and PLS. PCA was used to reveal spectral differences between meat samples, which helps in identifying the presence of pork in adulterated meat products. OPLS-DA was effective in discriminating between pure and adulterated meat samples with high accuracy. It can successfully identify both binary and ternary adulteration in meat products. In addition, PLS regression was successfully used for predicting the levels of pork adulteration. Moreover, several key markers and techniques were identified, such as methionine, glutamine, leucine, and isoleucine. Methionine and glutamine levels are higher in beef, making them useful markers for identifying pork adulteration in beef products. Meanwhile, leucine and isoleucine were also potential markers for detecting pork adulteration. Their presence and concentration can help differentiate between pure and adulterated meat samples (Leng et al., 2023).
Proton-NMR (1H-NMR) spectroscopy was also successfully used to differentiate meat species of raw meat and processed meat products consisting of beef, pork, lamb, and poultry. The analysis was assisted using chemometrics of PCA and LDA. The extraction of samples was based on aqueous extraction combined with ultrafiltration. The ultrafiltration was aimed to reduce macromolecular components in the extract (Çelebier, 2020). NMR measurement was performed in untargeted mode. A 99% correct prediction rate was achieved in predicting samples according to their species. Meanwhile, a slightly lower prediction rate (93%) was obtained in processed meat products. These results suggested that 1H-NMR spectroscopy in combination with chemometrics is promising for the differentiation of meat species further for halal authentication purposes (Decker et al., 2022).
NMR spectroscopy coupled with multivariate analysis PCA and OPLS-DA was also successfully used for the detection of the adulteration in chevon, chicken, donkey, and beef meats. A total of 37 metabolites was identified from the gluteal muscle tissues of chicken, goat, beef, and donkey. PCA could not completely differentiate samples, however, supervised pattern recognition of OPLS-DA demonstrated good separation and differentiation among each type of meat. The discriminating metabolites were investigated through the variable importance for projections. In addition, some metabolites including inosine, carnosine, carnitine, creatine, and acetate became the major discriminating metabolites to differentiate beef, donkey, and goat meat (Akhtar et al., 2021).
Conclusion
Analysis of non-halal meats either due to its animal origins (pork, canine, rat, wild boar) or due to slaughtering processes (zabiha and non-zabiha) is very important to ensure the halalness of meat products. The metabolomics approach in combination with chemometrics are reliable methods for differentiation between halal and non-halal meats as well as meats coming from zabiha and non-zabiha slaughtering processes. LC-MS/MS, two dimensional GC-MS and NMR spectroscopy are emerging analytical methods for metabolomics studies for analysis of non-halal meats. Indeed, these methods could be subjected to collaborative studies among competent laboratories to support the implementation of halal certification process.













