Introduction
Meat and meat products are significant sources of nutrition for humans, including proteins, lipids, minerals, and vitamins. Every year, beef consumption rises, and one consequence of this rise is the mixing of beef with other meats, such as pork, during processing. Furthermore, meat adulteration may violate religious beliefs; for example, Kosher and Halal food laws prohibit the consumption of pig or pork-related items (Alzeer et al., 2018; Lim and Ahmed, 2016). Many strategies have been implemented to ensure the authenticity of meat and meat products (Abbas et al., 2018; Mahbubi et al., 2019) but still the coverage is insufficient, and certification of all meat products for protection against adulteration is unfeasible. As a result, effective methods are necessary for assuring the meat industry’s proper development, and rapid, comprehensive, accurate, and reliable detection technologies are crucial to achieving this goal. One approach for guaranteeing food authenticity could be metabolomic and lipidomic technology (Ali et al., 2020b; El Sheikha et al., 2017; Islam et al., 2021; Vanany et al., 2020) compared to existing methods such as polymerase chain reaction (PCR), which has the limitation of being easily degraded in processed foods so that it has the potential to cause false negatives (Jannat et al., 2018; Jannat et al., 2020; Lubis et al., 2016). Metabolomic as method with high accuracy, comprehensive analysis of the whole metabolome which refer to the full complement of small molecule, while lipidomic explored area within lipid analytics and more specifically meat adulteration, so this method can used authentication of meat product (Emwas et al., 2019; Trivedi et al., 2016).
Metabolomics is a method of qualitative and quantitative analysis of metabolites in cells, tissues and biological fluids with small molecular weights of 100 to 1,000. Metabolites are the result of gene expression derives from the interaction between the genomic system and the environment (Crestani et al., 2020; Erban et al., 2019). Metabolites consist of an intermediate compound and metabolism product. The fingerprint characteristics of metabolites found in meat, such as amino acids, sugars, organic acids, nucleic acids, and their derivatives, could be provided through metabolomics. With minimal sample preparation, this approach can examine the components globally (De Paepe et al., 2018). The metabolomics approach to pork and beef study could provide a picture of the metabolites present in both foods. The metabolite profile of pork differs greatly from that of beef, hence the latter’s metabolite profile could be used as a baseline for determining the meat’s authenticity. Metabolomic and lipidomic can also distinguish pork mixture in mutton and chicken by looking at the different metabolites and lipid profiles (Wang et al., 2020). Lipidomics could be another way to look into the existence of pork mixture in meat products (Castro-Puyana et al., 2017; Yang et al., 2019).
Lipidomics is a comparatively new field of study, and it is developing quickly because to recent advancements in data analysis, bioinformatics data processing, and system biology techniques that are connected to other omics systems (Kliman et al., 2011). Various types of lipids, such as fatty acids and triglycerides, are found in the metabolome and the most distinctive biomarkers in which each type of tissue in meat has a different lipid profile, making it possible to identify unwanted species in a food product. For each animal species, there are a number of fatty acids located in specific tissue that can be used to differentiate between the various other animal species found in meat products. However, another significant benefit of lipidomic research is that it enables for the identification of animal species based on their lipid profiles (Ballin, 2010; Dettmer et al., 2007; Domínguez et al., 2019). Pork has a completely different lipid profile compare than beef. To evaluate whether a product contains pork or beef, the lipid profile of pork and beef can be utilized as a guideline. Liquid chromatography mass spectrometry (LC-MS) could be used in metabolomic and lipidomic techniques to investigate the authentication of meat.
The LC-MS method that integrates metabolomic and lipidomic analysis in pork and beef is a new technology with excellent sensitivity and provides the fingerprint of metabolites and lipids in biological samples. Organic chemicals and some inorganic substances can be analyzed using LC-MS (Gorrochategui et al., 2016). Sample preparation, data acquisition, and subsequent processing could be made easier using a mix of chromatographic and mass spectrometry techniques (Moosmang et al., 2019). This approach is frequently used to determine and characterize tiny molecules in meat products with high separation, such as metabolites and lipids. One of the advantages of LC-MS is the simplicity with which samples can be prepared. Mass spectrometry could verify that the types of metabolites and lipids present in the sample. Another benefit of LC-MS in metabolomic and lipidomic research is that it can identify all types of metabolites and lipids in a single sample run. This makes both analyses very efficient while using LC-MS (Neef et al., 2020).
Based on the preceding description, more investigation is necessary to answer the question of whether metabolite and lipid profiles can be utilized to determine the authentication validity of meat. To answer this question, a systematic review involving a comprehensive metabolomic and lipidomic approach is required, and it may be able to provide a comprehensive reference in the assessment of meat products (Demirhan et al., 2012; Mostafa, 2020; Pranata et al., 2021; Rohman and Che Man, 2011).
Analytical Methods for Authentication of Meat Products
Authenticity detection technologies for meat and meat products, such as PCR based on deoxyribonucleic acids (DNAs), protein technologies, and spectroscopic technologies based on specific metabolites, have all been developed in the previous two decades (Li et al., 2020). Presently, PCR (Amaral et al., 2017) and proteomics methods are routinely used for the species authentication (von Bargen et al., 2014). PCR is the most extensively used method for determining of meat products based on the presence of DNA, whereas proteomic is a method for determining of meat products based on their protein profile (Nakyinsige et al., 2012). Furthermore, PCR may detect pork DNA in a product (Izadpanah et al., 2017; Nakyinsige et al., 2012; Yuswan et al., 2018) and can detect a very small number of DNA copies. The hybridization of particular oligonucleotides to the target DNA and the synthesis of millions of copies flanked by these primers are the foundations of PCR amplification. Amplification of DNA fragments followed by agarose gel electrophoresis for fragment size verification is the most basic PCR approach for determining the presence of any species in meat products. Appropriate genetic markers are chosen to create the examination in order to properly detect species by PCR (Izadpanah et al., 2017). Porcine gelatin of pork can also be employed as an indicator of meat products in PCR analysis. The presence of DNA porcine gelatin of pork in the sample can be determined using the PCR technique (Rohman et al., 2020). The proteomic technique is another method for determining the validity of meat products. The goal is to determine the validity of meat products by examining for proteins, biological activity, post-translational modifications, and interactions in cells, as well as identifying the proteome in response to changes in porcine biological circumstances in the samples (Zamaratskaia and Li, 2017). Liquid chromatography quadrupole time of flight mass spectrometry (LC-QTOF-MS) is a method for determining the type of meat using a powerful tool for identifying protein peptides (Sarah et al., 2016; Zamaratskaia and Li, 2017). Protein extraction precedes mass spectrometry (MS) or LC-QTOF-MS analysis in the proteomic analysis method. In proteomics, mass spectrometry is the most typical approach for detecting proteins or peptides. This method has a wide range of applications, including meat science research, but it is hampered by the large biochemical heterogeneity of proteins and the inability to detect low protein levels. The detection of meat products using a proteomic technique has a high selectivity because only certain types of pork peptides can be found (Stachniuk et al., 2019). The PCR and proteomics methods both have their own set of difference when it comes to detecting adulteration in meat and meat products, Therefore, recent advances of the omics technologies (particularly metabolomics and lipidomics) are comprehensively discussed in this review.
Beef Meat and Its Products
Meat and its products are consumed widely throughout the world as a source of high-quality protein, essential amino acids, vitamins, and necessary minerals (Demirhan et al., 2012). A suitable analytical technique, such as headspace solid-phase micro extraction/gas chromatography-mass spectrometry, which employs volatile compounds to identify the meat authenticity, is used to ensure the authentication of beef and its products. The presence of alcohol compounds, 2-butanol and 1-octen-3-ol in a mixture of beef and pork can be used as a reference. These chemicals indicate presence of a pork mixture in meat products (Hossain et al., 2020; Pavlidis et al., 2019). Another method to ensure the meat authenticity is by EvaGreen real-time PCR. This validated method is able to detect pork DNA specifically in meat product samples. This method can detect 0.01%–100% pork contamination in beef meatballs with high accuracy and precision. In addition to the two procedures mentioned above, other methods such as Fourier-transform infrared spectroscopy (FTIR) and LC-MS can be used to determine the authenticity of beef and its products (Lubis et al., 2016; Yuswan et al., 2018). FTIR also can be used to assess the meat authenticity. This approach can identify functional groups in proteins as pig identifiers, allowing pork-containing products to be recognized (Lubis et al., 2016). By looking at the peptide fingerprints found in pork, LC-MS can be utilized to detect the meat authenticity. Moreover, chemometric technique also used to assess the type of peptide present in pork using this peptide fingerprint. Pork marker peptide is the result of this chemometric analysis, and it is utilized to determine the meat authenticity (Yuswan et al., 2018).
Chicken Meat and Its Products
Chicken meat is one of the meat products that provide the body with essential amino acids, fatty acids, and vitamins that the body need (Ali et al., 2019). The analytical methods are needed to verify that chicken meat authenticity. The high-sensitivity technology for detecting the presence of a pork mixture in the product is necessary in this case. LC-QTOF-MS/MS is the analytical method for analyzing chicken meat and its products. The protein acquired from the MS spectra is matched to the absolute protein expression database using the proteomic principle. The authenticity of chicken meat products can be determined using a peptide derived from other meat such as pork (Montowska and Fornal, 2017). Another method to examine the authenticity of chicken meat and its products is proteomic analysis using matrix-assisted laser desorption/ionization-time of flight. Determination of halal chicken meat is not only based on the presence of a mixture of pork but also the method of slaughter. In this method, it is possible to detect the proteome of chicken meat slaughtered in a halal and non-halal way. Beta-enolase, pyruvate kinase, and creatine kinase compounds are the ones that could have higher levels when slaughtered in an illegal manner (Salwani et al., 2015).
Animal Fat Products
There is a growing need for animal fat nowadays, based on the Statistic Government Bureau in Indonesia, export value of animal and vegetable oil fats has increased from 19,329 million USD in 2018 to 197,095 million USD in 2020 (Qodri, 2018). Animal fat is essential product that has various purposes in the body, including providing energy and forming adipose tissue. Fat is the most energy-dense food, pro-ducing 9 kcal per gram, 2.5 times the energy provided by carbohydrates and protein in the same quantity. Fat can produce fatty acids and cholesterol needed to form cell membranes in all organs.
Halal meat products are important since consumption of halal meat might influence one’s attitude towards halal slaughter (Jalil et al., 2018). To be declared halal, meat products must meet a number of conditions relating to their preparation, condition for analysis such as towing before analysis, and composition. Consumers may trust halal meat labeling to ensure that the meat is of good quality, high value, safe, animal-friendly, and environmentally friendly (Haleem et al., 2021; Lim et al., 2022). Animal fats such as chicken and beef fat are permissible (Sin and Sin, 2019), however pork fat is prohibited for Moslem according to Shariah (Islamic law; Ahmad et al., 2018).
Beef fat is one type of meat products that is usually consumed nowadays. Beef is abundant in fat and contains important nutrients such as essential fatty acids, in addition to protein. Consumers’ concerns and awareness about the eating of high-fat meat items have an impact on meat consumption patterns (Mahbubi et al., 2019). The following is a representation of beef fat content: Saturated fatty acids, n-6 polyunsaturated fatty acids, n-3 polyunsaturated fatty acids, and trans fatty acids are the different types of fatty acids. Fatty acids in beef vary based on genotype, muscle type, and feeding methods in general. Long-chain n-3 and n-6 polyunsaturated fatty acids found in beef provide extra health benefits, including improved maternal and child health, growth and development, and cognitive function and psychological state in humans (Troy et al., 2016). Animal fat can be investigated from the use of technology including spectroscopy and chromatography (Rohman and Fadzillah, 2021). Raman spectroscopy is one of the spectrophotometric methods that can be used to identify the authenticity of animal fats (Lee et al., 2018). The resulting spectra could be forwarded using various types of databases in this manner, allowing them to determine the types of unsaturated and saturated fatty acids. The types of fat are not only qualitatively but also quantitatively examined in this method. It could be possible to accurately detect the type and amount of pork animal fat. Another method of authenticity analysis of animal fat by chromatographic method is using high-performance liquid chromatography nuclear magnetic resonance (HPLC-NMR) and gas chromatography–mass spectrometry (GC-MS). HPLC-NMR is able to separate well the lard compounds in the sample which is then followed by reading the structure of the compound. Pork fat has the unique characteristic of containing polyunsaturated fatty acids. This fat could be used as a target to determine the authenticity of animal fat. Determination of the authenticity of animal fats can also be conducted with a targeted metabolomic approach using GC-MS, a simple method with good separation technique (Fadzillah et al., 2017; Heidari et al., 2020). Methyl myristate, methyl palmitate, methyl oleate, and methyl stearate are examples of targeted metabolites of lard that can be analyzed in the samples.
Authentication of Meat Product Using Metabolomics and Lipidomics Approaches
It is critical to determine authentication of meat. In order to acquire valid results that may be used to declare authenticity, new methods are still being developed. By examining the metabolite profile, one way for determining the authenticity of meat and its products is metabolomics. Metabolomic studies can use either spectrophotometry or chromatography (Muroya et al., 2020). The spectrophotometry used in metabolomic analysis is Ultraviolet-Visible, Infrared, Raman, and nuclear magnetic resonance (NMR) combined with chemometrics for spectral data. MS (Junot et al., 2014) and non-MS such as NMR are the most widely used methods (Consonni and Cagliani, 2019). In addition, different types of separation techniques are incorporated in most MS-based, depending on the lipophilicity and polarity of the desired metabolite. Combined with statistical analysis, multivariate analysis, and bioinformatics databases, metabolomics provides for finding biomarkers (Sugimoto et al., 2012).
The most widely utilized multivariate analyses in certified meat products are principal component analysis (PCA), partial least squares-discriminant analysis (PLS-DA), and orthogonal projections to latent structures discriminant analysis (OPLS-DA; Dailey, 2017). PCA is used to narrow down the list of metabolites and lipids to only the most important ones (Zhang et al., 2022). Other animal species that are present in meat can be identified by PCA when validating LC-MS data (Kang et al., 2022). Typically, Minitab, Orange, R Studio, and Unscamble are used for PCA analysis. The PLS-DA classification is a good choice for recognizing meat products. This is because the PLS-DA approach, whose implementation is exceedingly user-friendly and is extensively utilized in the most well-known statistical software packages like R Studio, is also quite popular. Additionally, PLS-capacity DA’s to analyze highly linear and noisy data is one of its advantages (Utpott et al., 2022).
Using R Studio software, OPLS-DA provides a quick, easy, and effective multivariate analysis. In order to find meat-specific quantitative peptides created using liquid chromatography-tandem mass spectrometry (LC-MS/MS), OPLS-DA was applied to halal analysis. To choose species-specific peptides that significantly assist in classification, the OPLS-DA model was developed. Three distinct quantitative peptides were found in products with various beef proportions after the statistical process flow. LC-MS/MS was used to build quantitative methodologies for the specific quantitative peptides selected. Commercial beef products were subjected to quantitative results. The devised method is extremely precise, repeatable, and sensitive. According to Kang et al. (2022), LC-MS/MS integration with OPLS-DA is an efficient method to screen for particular quantitative peptides and certify beef product.
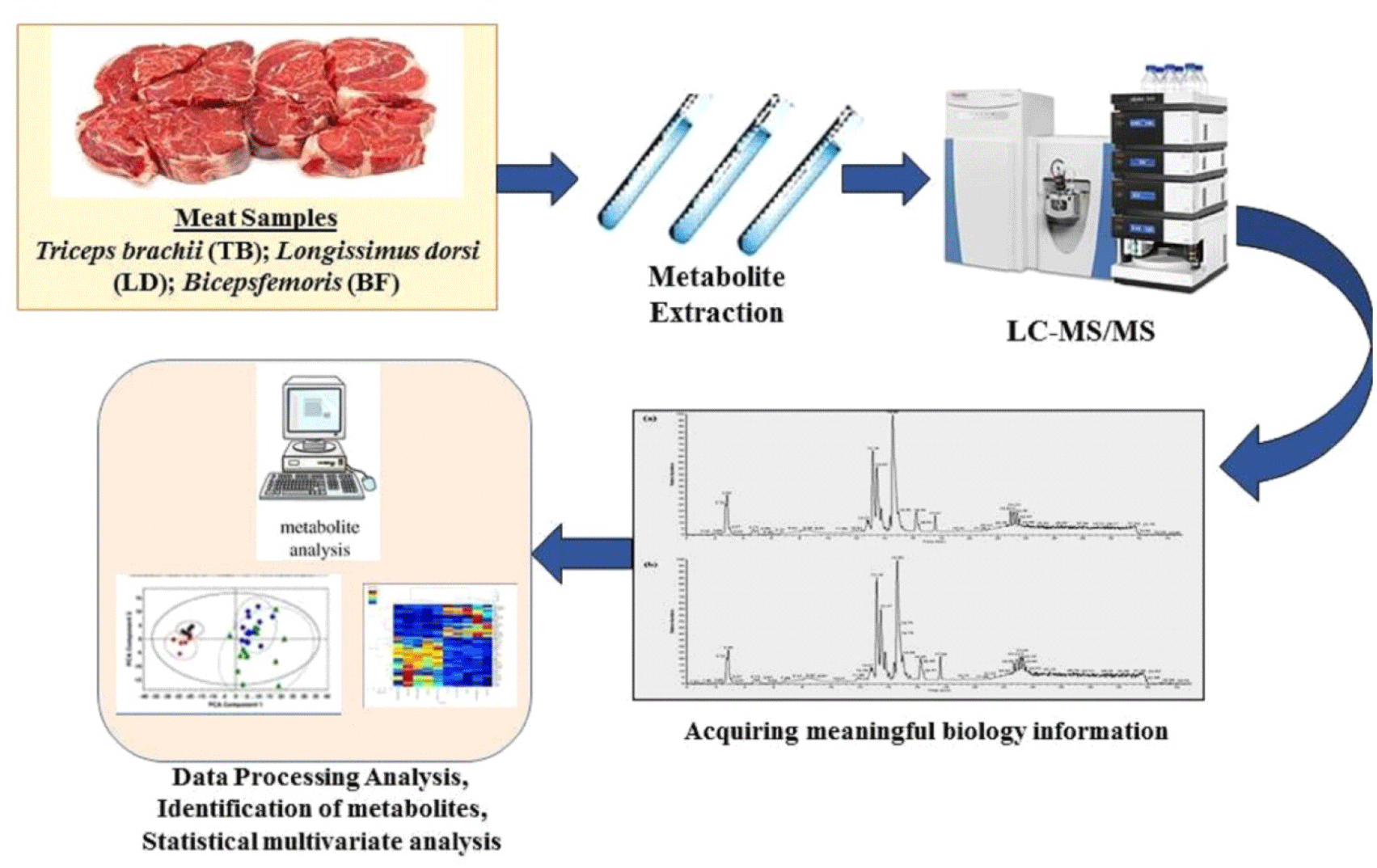
Apart from spectrophotometry, chromatography is also used in metabolic analysis. In metabolomic analysis, the use of LC-MS is critical, and one example is the use of ultra-high performance liquid chromatography quadrupole time-of-flight with rapid evaporative ionization mass spectrometry, which can provide metabolites in meat (Wang et al., 2020). The workflow of metabolomics analysis in meat samples can be seen in Fig. 1. Besides metabolomics, lipidomics is another way for determining the validity of meat products. The result of metabolite analysis can be used for authentication meat products such as myosin-2 (Yuswan et al., 2018), 3-Oxohexane acid glycerides, arabitol, creatinine, glycine and phosphate (Trivedi et al., 2016). Also the lipid component that can be used for authentication such as sphingomyelins, cerebrosides, globosides, gangliosides or sulfatides (Trivedi et al., 2016).
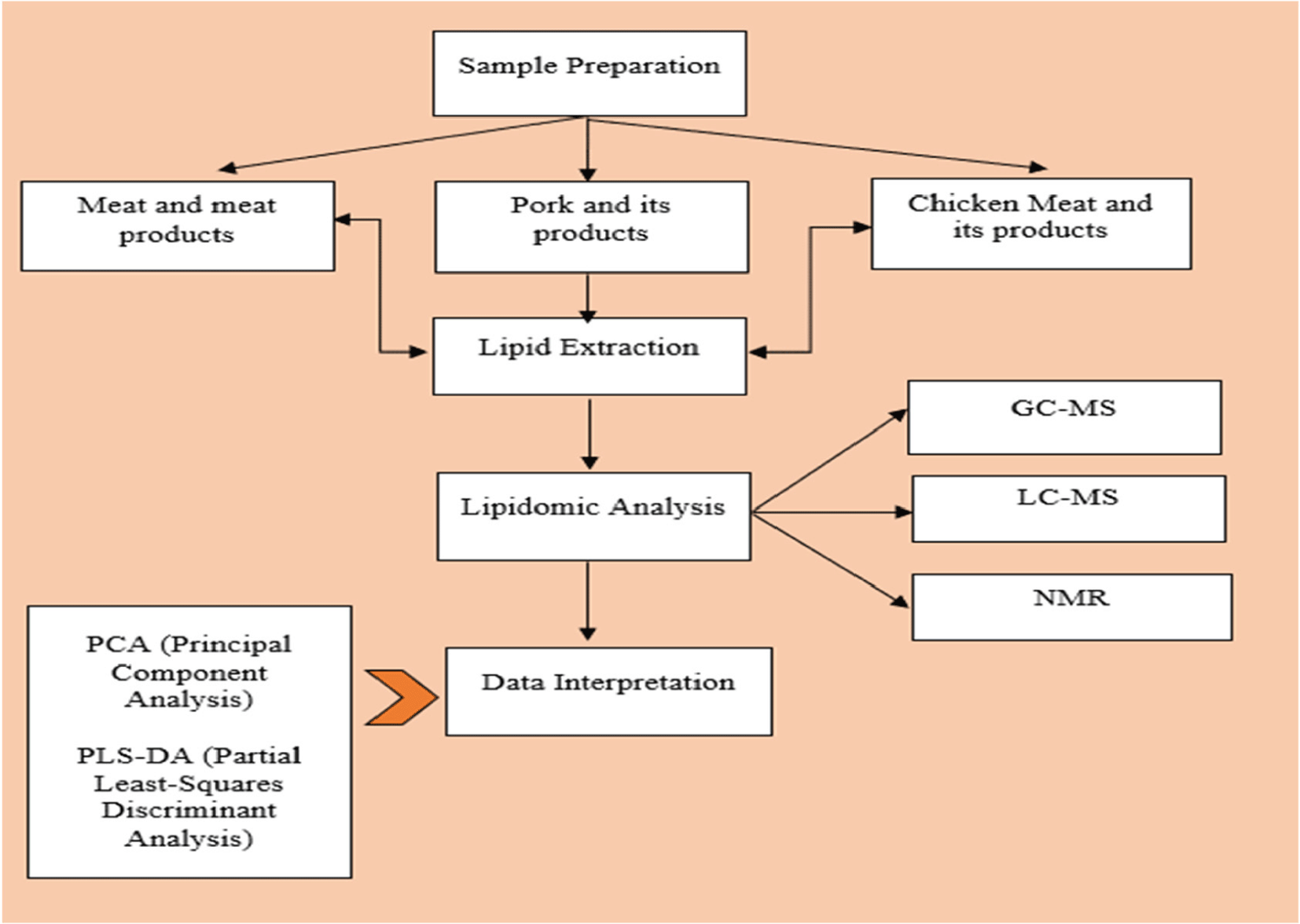
The workflow summarizing the different steps in lipidomics analysis can be seen in Fig. 2. The study of lipid profiles that can be applied to meat products in order to assess authenticity is known as lipidomics. Pork’s lipid profile is undoubtedly different from that of other meats, and this unique lipid profile can be used to confirm the meat authenticity. Lipidomic analysis to determine meat authenticity can use LC-MS, one of which uses liquid chromatography electrospray ionization tandem mass spectrometric. This method is able to interpret the lipid profile combined with MS data. The lipid profile of pork-containing meat products can be identified well (Dirong et al., 2021).
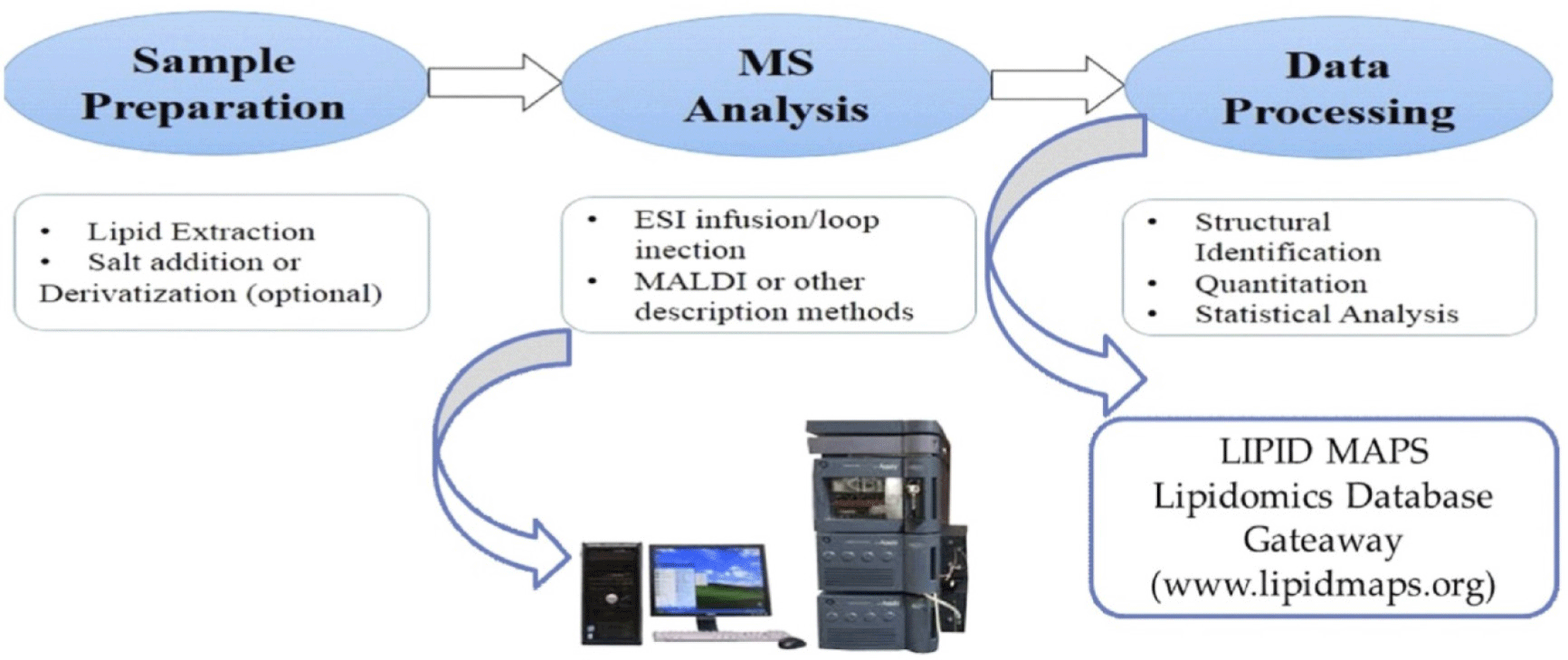
The information on the compounds resulting from LC-MS in lipidomic and metabolomic study is enormous, reaching in the thousands. The metabolites that have been detected are then determined for the identified compounds using the databased compound discover, and the profile of lipids and sub-lipids is determined using the databased LipidSearch (Korf et al., 2019; Medema and Fischbach, 2015). Due to the size of the database’s results for metabolites and lipids, multivariate analysis is necessary to identify the metabolites and lipids that will serve as meat products’ authenticity indicators (Trivedi et al., 2016).
Metabolomic Studies
Metabolomics is the study of the profile of low molecular weight metabolites. The metabolomics approach can be applied to determine the authenticity of meat. The metabolite profile in pork could certainly be different from that of beef or other meats. For example, research by Rocchetti et al. (2020) was able to identify metabolites in pigs with ultra-high performance liquid chromatography-quadrupole time-of-flight mass spectrometry (UHPLC-QTOF-MS) which obtained hexanoylcarnitine, 4-hydroxy-2-nonenal, 6-hydroxypentadecanedioic acid, 9S, 11S, 15S, 20-tetrahydroxy-5Z, 13E-prostadienoic acid (20-hydroxy-PGF2a), sativa acid, and glycerophospholipid. Moreover, Ali et al. (2020a) was able to identify glucose, amino acid, inosine, hypoxanthine, and arginine in broiler chickens slaughtered in an illegal manner using UHPLC-QTOF-MS. Furthermore, Jia et al. (2021) mentioned that 103 metabolites could be identified such as L-phenylalanine, L-isoleucine, L-histidine, guanosine, guanine, creatinine, glutathione, and nicotinic acid in goat meat using UHPLC-QTOF-MS. Several studies have reported the use of highly accurate metabolomic technologies to evaluate metabolite profiles, indicating that an approach based on this method could be used to determine qualities in the meat.
Metabolomics is a method for determining the numerous metabolite profile found in pork and beef. The qualities of the metabolite profiles in pork and beef could be distinguished by using these metabolites. Chromatography in mass spectrometry is used to separate metabolites from samples. Ultra performance liquid chromatography (UPLC) is usually being used to separate metabolites because of its ability to separate well. The time of flight-mass spectrometry apparatus could be coupled to the UPLC device, making it easier to get the research data. The ultra performance liquid chromatography-quadrupole time-of-flight mass spectrometry (UPLC-TOF-MS) for metabolomic analysis was also described by Jia et al. (2021) and identified 103 metabolite profiles in goat meat. The UPLC-TOF-MS method is an effective way to figure out the metabolite profiles of meat samples. The data could be compared to the Compound Discovered database, and multivariate statistics could be used to interpret the results. Pork’s metabolite profile, which distinguishes it from beef, could be utilized as a standard for determining authenticity in food samples. Metabolites such as decanoylcholine, glycyl-lysine, and oleic acid can be used to authenticate meat products. The resume authentication of meat product using metabolomics can be shown in Table 1. Moreover, the metabolite extraction method was according to Jang et al. (2019), sample was mixed with 150 μL methanol: acetonitrile: water (40: 40: 20, extraction solvent), vortexed, and immediately centrifuged at 16,000×g for 10 min at 4°C. The supernatant was collected for analysis.
| No | Title | Refs | yr | Objectives | Equipment | Metabolite results |
|---|---|---|---|---|---|---|
| 1 | 1H-NMR-based metabolomic profiling and taste of stewed pork-hock in soy sauce | (Yang et al., 2019) | 2019 | Steward pork | 1H-NMR | Amino acids, sucrose, β-glucose, acetate, and creatinine |
| 2 | LC–QTOF-MS identification of porcine- specific peptide in heat treated pork identifies candidate markers for meat species determination | (Sarah et al., 2016) | 2016 | Meat (pork, beef, chicken, and chevon) | LC-QTOF-MS | Seven porcine-specific peptides, two were derived from lactate dehydrogenase, one from creatine kinase, and four from serum albumin protein |
| 3 | A volatilomics approach for off-line discrimination of minced beef and pork meat and their admixture using HS-SPME GC/MS in tandem with multivariate data analysis | (Pavlidis et al., 2019) | 2019 | Meat (beef and pork) | GC-MS | Alcohols, 2-butanol, and 1-octen-3-ol |
| 4 | Chemometrics-assisted shotgun proteomics for establishment of potential peptide markers of non-halal pork (Sus scrofa) among halal beef and chicken | (Yuswan et al., 2018) | 2018 | Meat (beef, chicken, and pork) | LC-MS | 7 Peptides marker |
| 5 | Discrimination between vegetable oil and animal fat by a metabolomics approach using gas chromatography-mass spectrometry combined with chemometrics | (Heidari et al., 2020) | 2020 | Animal fats | GC-MS | Methyl myristate, methyl palmitate, methyl oleate, and methyl stearate |
| 6 | Liquid chromatography quadrupole time-of-flight mass spectrometry and rapid evaporative ionization mass spectrometry were used to develop a lamb authentication method: A preliminary study | (Wang et al., 2020) | 2020 | Meat | UHPLC-QTOF-MS | 42 Potential metabolites |
| 7 | Impact of a pitanga leaf extract to prevent lipid oxidation processes during shelf life of packaged pork burgers: An untargeted metabolomic approach | (Rocchetti et al., 2020) | 2020 | Meat | UHPLC-QTOF-MS | Hexanoylcarnitine, 4-hydroxy-2-nonenal, 6-hydroxypentadecanedioic acid, 9S,11S,15S,20-tetrahydroxy-5Z,13E-prostadienoic acid (20-hydroxy-PGF2a), sativic acid |
| 8 | Effect of different slaughtering method on metabolites of broiler chickens using ultra-high performance liquid chromatography-time of flight-mass spectrometry (UHPLC-TOF-MS) | (Ali et al., 2020a) | 2019 | Chicken meat | UHPLC-QTOF-MS | Histidin, inosin, hypoxantine |
| 9 | Meat, the metabolites: An integrated metabolite profiling and lipidomics approach for the detection of the adulteration of beef with pork | (Trivedi et al., 2016) | 2016 | Meat | GC-MS and UHPLC-MS | Arabitol, citric acid, glucose 6-phosphat, glycine, malic acid |
1H-NMR, hydrogen-1 nuclear magnetic resonance; LC-QTOF-MS, liquid chromatography quadrupole time of flight mass spectrometry; HS-SPME, headspace solid-phase micro extraction; GC-MS, gas chromatography-mass spectrometry; LC-MS, liquid chromatography-mass spectrometry; UHPLC-QTOF-MS, ultra-high performance liquid chromatography-quadrupole time-of-flight mass spectrometry; UHPLC-TOF-MS, ultra-high performance liquid chromatography-time-of-flight-mass spectrometry; UHPLC-MS, ultra-high performance liquid chromatography-mass spectrometry.
Lipidomic Studies
Lipidomics is a novel branch of research that examines the structure and function of lipids produced in plant and animal cells, as well as their interactions with other lipids, metabolites, and proteins (Li et al., 2017) can be regarded as a relatively unexplored area in food analysis (Aiello et al., 2011) and more specifically on adulteration of meat. Using the LC-MS approach, Mi et al. (2019) were able to successfully detect the component of lipids in several species of pork in China. They results showed that 61 types of glycerolipids, 17 glycerol phospholipids, 4 sterol lipids, 2 sphingolipids, 3 polyketides, 7 fatty acids, and 6 phenol lipids. Trivedi et al. (2016) used GC-MS and UHPLC-MS to evaluate lipidomic profiles of beef contaminated with pork. They found lipid components in beef, such as sphingomyelins, cerebrosides, globosides, gangliosides or sulfatides. Furthermore, Artegoitia et al. (2019) investigated the metabolomic and lipidomic profiles of beef fed efficiency feed using the UPLC-QTOF-MS technique, the results showed there were 20 types of phospholipids and cholesterol, such as phosphatidylcholine (PC), phosphatidilethanolamine, lysophosphatidilcholine, and lysophosphatidyleththanolamine. These studies have proven that the lipidomic method may be used to examine lipids in pork and beef utilizing chromatographic techniques to separate lipids for further examination. The type of chromatography used in previous study was column chromatography. With a single run, column chromatography may separate a large number of organic molecules in a short amount of time. Several scientific publications have reported the use of very accurate lipidomic technologies to examine lipid profiles, indicating that this technology could be used to determine meat authenticity, does not containing other meat such as pork. Glyserolipid and spingolipid are examples of lipids that can be used to authenticate meat.
In addition, the sample pretreatment for lipid and metabolite extractions are using similar methods in a different mixture. The lipid from meat were extracted according to the method of Harlina et al. (2021), whereas, meat sample were homogenizer in a mixture of chloroform: methanol: distilled water (120:120:60, v/v/v) at 11,000 g using a homogenizer for 2 min. Then, the homogenized mixture was treated with ultrasound (20°C, 80% power, 30 min). The mixture was filtered through a Buncher filter funnel. The chloroform phase (bottom phase) was drained off into an Erlenmeyer flask. The lipid in chloroform was decanted into a round-bottom flask through a filter paper. Before it was evaporated at 55°C using rotary evaporator and the residual solvent were removed by flushing with nitrogen. The lipid was stored at –20°C until was analyzed.
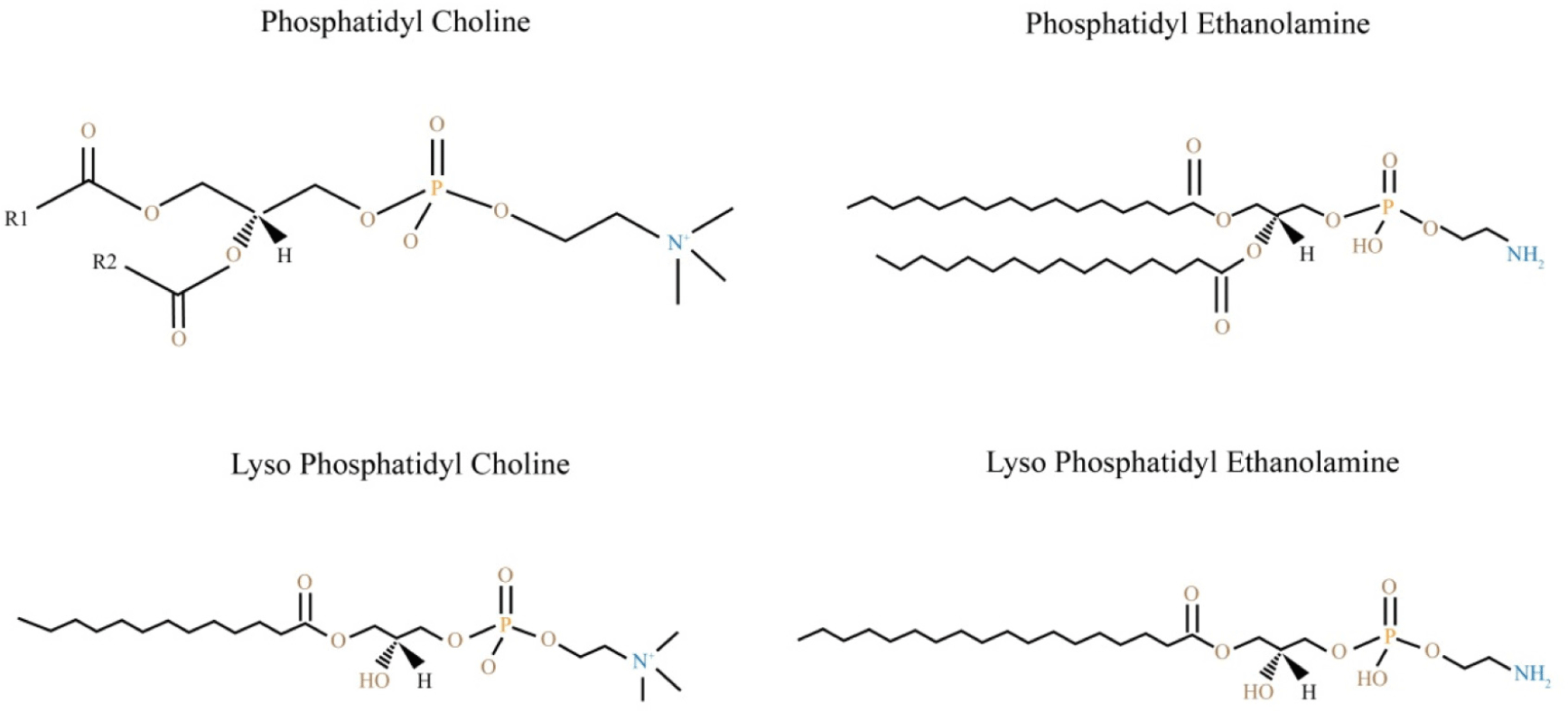
Primary lipids such as cholesterol and its esters, as well as triglycerides, are found in nonpolar lipids, lipids are compounds that are soluble in nonpolar solvent such as chloroform (Han and Gross, 2005; Yu et al., 2020). Phospholipids, sphingolipids, rhamnolipids, and glycolipids are the most common lipid classes found in polar lipids. Furthermore, phospholipids are divided into numerous groups based on the phosphate classes, including such as PC, phosphatidylethanolamine (PE), phosphatidylinositol (PI), phosphatidylglycerol, phosphatidylserine (PS), and phosphatidic acid (Li et al., 2017). Glycerophospholipid categories in the meat can be seen in Fig. 3. PC, PE, PI, and PS are the major glycerophospholipids found in the membrane. They have various functions in the plasma membrane’s exoplasmic and cytoplasmic functions, and they provide a semi-permeable barrier to keep the cell intact (Arish et al., 2015).
Lipidomics is a technique for determining the numerous lipid species found in the food samples. According to Harlina et al. (2021), depending on the lipid species and head groups, lipids can be identified using positive or negative ions. The majority of phospholipids are found in both positive and negative ion modes as distinct adducts, including +H+, +NH4+ in positive mode and –H, +CH3COO, or +HCOO– in negative mode. Triglycerides and diglycerides are examples of neutral lipids that are all recognized in positive mode as NH4 adducts. However, only negative ion mode can identify the fatty acid contents of phospholipids, while positive ion mode can declare head group and/or neutral loss. Therefore, lipids are amphiphilic substances that ionize in both positive and negative modes. The MS based shotgun of lipidomics can be seen in Fig. 4.
Resume authentication of meat product using lipidomic can be shown in Table 2. UPLC is currently used to separate lipids since it can separate up to the sub-class of lipids. Q-extractive heated electrospray ionization can be coupled to the UPLC equipment, making it easier to get research data. Narváez-Rivas and Zhang (2016) described the use of the ultra performance liquid chromatography q-extractive heated electrospray ionization (UPLC-QE-HESI) for lipidomic analysis. This method has good selectivity and accuracy for determination lipids in sample (Narváez-Rivas and Zhang, 2016). They used this equipment to detect 430 lipid profiles in plasma. The UPLC-QE-HESI is an excellent tool for determining the lipid profiles of meat samples. The research data can be compared to lipid databases evaluated using multivariate statistics. Pork’s lipid profile, which distinguishes it from beef, could be utilized to determine authenticity in food sample (Holčapek et al., 2018; Narváez-Rivas and Zhang, 2016). The advantages and disadvantages of metabolomic and lipidomic for authentication in meat product can be seen in Table 3.
| No | Title | Refs | yr | Objectives | Equipment | Lipids result |
|---|---|---|---|---|---|---|
| 1 | Authentication of butter from lard adulteration using high-resolution of nuclear magnetic resonance spectroscopy and high-performance liquid chromatography | (Fadzillah et al., 2017) | 2017 | Lard | NMR, HPLC | Triacylglycerol and fatty acids |
| 2 | Quantitative analysis of lard in animal fat mixture using visible Raman spectroscopy | (Lee et al., 2018) | 2018 | Lard | Raman spectroscopy | Quantitative fat oil |
| 3 | Liquid chromatography quadrupole time-of-flight mass spectrometry and rapid evaporative ionization mass spectrometry were used to develop a lamb authentication method: A preliminary study | (Wang et al., 2020) | 2020 | Meat | UHPLC-QTOF-MS | Multiple triglyceride (TG), diacylglycerol (DG), and PL |
| 4 | Characterization and discrimination of selected China’s domestic pork using an LC-MS-based lipidomic approach | (Mi et al., 2019) | 2019 | Raw pork meat | LC-MS | 61 glycerolipids, 17 glycerophospholipids, 4 sterol lipids, 2 sphingolipids, 3 polyketides, 7 fatty acyls and 6 prenol lipids |
| 5 | Meat, the metabolites: An integrated metabolite profiling and lipidomics approach for the detection of the adulteration of beef with pork | (Trivedi et al., 2016) | 2016 | Meat | GC-MS and UHPLC-MS | Fatty acid |
NMR, nuclear magnetic resonance; HPLC, high performance liquid chromatography; UHPLC-QTOF-MS, ultra-high performance liquid chromatography- quadrupole time-of-flight mass spectrometry; LC-MS, liquid chromatography-mass spectrometry; GC-MS, gas chromatography-mass spectrometry; UHPLC-MS, ultra-high performance liquid chromatography-mass spectrometry.
| No | Method | Advantages | Disadvantages | Refs |
|---|---|---|---|---|
| 1 | Metabolomic | Has a high accuracy value, Comprehensive analysis of the entire metabolome associated with the complete complement of small molecule, have been used to analyze organic components. | Requires proper instruments for analytical processes such as LC-MS, Metabolomic analysis equipment is expensive. | (Emwas et al., 2019; Trivedi et al., 2016) |
| 2 | Lipidomic | Areas explored in food analysis and more specifically meat adulteration, can be used detection meat with quickly. | The data obtained are limited to lipid compounds and sub lipids. | (Trivedi et al., 2016) |
Future Perspective
The techniques of metabolomics and lipidomics can be used to identify meat products. The combination of the two will give a clearer picture of the meat profile (Wang et al., 2021a; Wang et al., 2021b). The complete lipid and metabolite profiles can effectively distinguish between meat varieties (Wang et al., 2021a; Wang et al., 2021b; Wu et al., 2021; Zhang et al., 2021). In the identification of meat products, combining metabolomic and lipidomic approaches could provide a more comprehensive overview (Ellis et al., 2016; Munekata et al., 2021). The combination of these two methods can be used to determine the authentication of meat products, by looking at the metabolites and lipids in meat products whether they contain pork (D’Alessandro and Zolla, 2013). In comparison to the metabolomic or lipidomic method alone, the combination of these two procedures could precisely evaluate the authentication of meat products (Capozzi et al., 2017; Chin et al., 2009; Picó, 2015; Yuliana et al., 2022).
Conclusion
Metabolomics is the study of metabolite profiles that can be used to identify the authenticity of meat products by examining metabolite profiles in pork that are not represented by beef. Lipidomics is a lipid profile analysis that may be used to determine the authenticity of meat products by examining the lipid profile of pork, which beef excludes. LC-MS instruments such as the UHPLC-QTOF-MS and UPLC-QE-HESI, which are directly connected to the metabolite and lipid databases software, can be used for metabolomic and lipidomic approaches to assess the authenticity of meat products. Combination metabolomic and lipidomic approaches to assess the authenticity of meat products would be more accurate because it can describe extensive lipid and metabolite profiles in meat.