Introduction
Artisanal cheeses are passed on from one generation to another and produced in a particular way that is unique to a certain local area, region, or country with little or no processing. They are generally known for their diverse microbiota, potent sensory properties, small risks, and some benefits. There are various question marks about artisanal cheeses considering their exposure to incomplete heat-treatment and their being made without a starter culture and their ambiguous microbial composition, all of which might result in a potential for high pathogen risks. Aside from all the pros and cons, demand for artisanal cheese increases progressively and traditional manufacturers try to adapt their production conditions and methods to vat milk with each passing day. In this aspect, determining the microbial community of artisanal cheeses is the key to enabling a safer and more beneficial cheese production by preserving the manufacturing practices, characteristic flavor, aroma, and texture of the product for many years. Hence, numerous studies have been executed on microbial diversity and dynamics of artisanal cheeses and possible microbial interactions during ripening over the last few years (Demirci et al., 2021; Dimov et al., 2021; Dugat-Bony et al., 2016; Onmaz et al., 2021). As a result of this interest, novel molecular techniques, such as 16S/ITS-based high-throughput sequencing, are frequently used to detect the microbiota of regional cheeses and their ripening roles worldwide (Dimov et al., 2021; Murugesan et al., 2018).
In Turkey, there are over 150 cheese varieties displaying an amazing diversity of organoleptic and textural properties. While most of them are produced in small quantities to meet local cheese requirements, some of them are commercialized throughout the country such as Ezine cheese, Tulum cheese, and Kuflu cheese. Kuflu cheese is a mold-ripened variety which is consumed in many rural and urban areas of Turkey. Despite the lack of clear historical records, Kuflu cheese manufacturing has a long story in Turkey and is the most typical artisanal cheese in the Central Anatolia Region (mainly in Konya, Karaman provinces). Such cheeses are made from skimmed or semi-skimmed unpasteurized ewe’s milk or its mixture with goat’s and cow’s milk in small quantities without any starter microorganisms, and these cheeses are usually characterized by blue-green molds that spontaneously grow on their surface.
With regards to the bacterial or fungal composition of artisanal Kuflu cheese or other mold-ripened Turkish varieties, some researchers have executed important studies using culture-dependent methods (Seri and Metin, 2021) and culturomics (Onmaz et al., 2021), however, to the best of our knowledge there is no metagenomic study of the microbiota of such cheese. As a matter of fact, metagenomic next generation sequencing has a superior capability to characterize the conserved and variable regions of the bacterial 16S or eukaryotic rRNA genes for the purpose of taxonomic classification. Normally, both bacteria and mold starter cultures are used to produce mold-ripened cheeses, otherwise, limited contamination of the microbiota is expected. However, the Kuflu cheeses examined in this study were traditionally produced under completely uncontrolled conditions. It is thought that the detection of pathogenic bacteria, intestinal commensals, and various contaminants is important for the food safety of Kuflu cheese, as well as an important literature information for artisanal mold-ripened cheeses. The determination of beneficial bacteria and yeasts is also valuable in terms of the health benefits that can be attributed to this type of cheese. For this reason, the cheeses were not produced by us and random Kuflu cheese samples that have a controversial place in terms of consumers were obtained from the bazaars. In this manner, it is essential to take metagenomic approaches to characterize the bacterial, yeast, and filamentous mold diversities and discuss their presence in Kuflu cheeses. In the present study, both bacterial and fungal biota in ten artisanal mold-ripened Kuflu cheese samples produced in Konya (Turkey) were characterized by high-throughput 16S DNA and ITS sequencing. The richness and distribution uniformity of both bacterial and fungal communities identified in Kuflu cheeses were determined by alpha diversity indices (Shannon, Simpson) and similarity/dissimilarity of microbial communities of cheeses was also detected by beta diversity indices [principal coordinate analysis (PCoA)].
Materials and Methods
A total of ten homemade artisanal Kuflu cheeses were supplied from ten different local producers of Konya province. From these samples, the first Kuflu cheese (MC1) was made from only raw cow’s milk, the second (MC2) from only raw goat’s milk and all the other cheeses were produced from the mixture of raw ewe’s (about 20%) and raw cow’s milk (about 80%). The samples were collected in October 2021 and cheese producers declared that they put these cheeses for sale after they have matured for 3–4 months. Cheese samples were quickly transferred to the laboratory and kept at –20°C until DNA extraction. The images of Kuflu cheeses analyzed in this work are presented in Fig. 1.
The pH values of cheese samples were analyzed by a calibrated pH-meter (Starter 3100 model, Ohaus, Parsippany, NJ, USA). The titratable acidity of samples was determined according to the titration method using 0.1 N NaOH (AOAC, 2016) and expressed as g lactic acid/100 g cheese. As for fat content, it was analyzed by Gerber method (Kleyn et al., 2019). Salt contents of samples were detected using the AgNO3 titration method suggested by Hayaloglu et al. (2005) and the results were provided as the salt content (%) in the dry matter. In addition to these, moisture content was detected by the gravimetric method (AOAC, 2016).
For extraction of total DNA directly from Kuflu cheese samples, the GeneMATRIX Bacterial DNA Purification Kit (EURX, Molecular Biology Products, Toronto, ON, Canada) was utilized based on the kit manufacturer’s instructions. Samples were taken from different points of the cheeses to represent the whole cheese.
The V3-V4 hyper-variable regions of the 16S rRNA gene were used for the examination of bacterial composition in Kuflu cheese samples (Demirci et al., 2022). (5’): TCG TCG GCA GCG TCA GAT GTG TAT AAG AGA CAG CCT ACG GGN GGC WGC AG and (5’): GTC TCG TGG GCT CGG AGA TGT GTA TAA GAG ACA GGA CTA CHV GGG TAT CTA ATC C were used as forward and reverse primers, respectively. After amplification with specific primers, purification was performed. At the index PCR step, the Nextera XT index kit (FC-131-1001/FC-131-1002) specific to Illumina technology was used. Library preparation and sequencing were performed using the NovaSeq Reagent kit according to the Illumina 16S metagenomic sequencing library protocol based on the yielding 2×250-bp (paired-end reads).
Fungi community was characterized by sequencing the ITS2 regions using specific ITS2 forward primer (5’): TCG TCG GCA GCG TCA GAT GTG TAT AAG AGA CAG GCA TCG ATG AAG AAC GCA GC and reverse primer (5’): GTC TCG TGG GCT CGG AGA TGT GTA TAA GAG ACA GTC CTC CGC TTA TTG ATA TGC. For this purpose, 15 ng of genomic DNA was employed. The ITS2 gene was amplified with specific primers. Indexing was constructed in the same way as for the 16S rRNA gene. AMPure beads (Beckman Coulter Genomics, Beckman Coulter, Brea, CA, USA) were used for the purification of the resulting libraries. For a more sensitive determination of library quantification, Qubit™ 4 fluorometer was used. Library preparation and sequencing were performed using the NovaSeq Reagent kit according to the Illumina ITS2 metagenomic sequencing library protocol (yielding 2×250-bp) on Illumina NovaSeq 6000.
After sequencing, the obtained data were converted to FASTA format for bioinformatic analysis. At first base, reads, primer, and barcodes with Phred score lower than 20 were trimmed. Data were analyzed using QIIME 1.9.1 (Caporaso et al., 2010) with a pipeline reported by Berni Canani et al. (2017) for taxonomic classification. After the evaluation of the read quality, similar sequences were clustered to detect the taxonomy of each sample. The Operational Taxonomic Units (OTU) were identified. OTUs representing less than 0.001% were excluded from the evaluation. To understand the microbial composition in the samples, the identified microorganisms were converted to percentages in each sample. The major OTUs in phylum- and genus-level were filtered at 1% and the stacked column charts were constructed using MS Excel. Shannon’s diversity and Simpson’s diversity indexes were used to calculate rarefaction curves (Willis, 2019). The PCoA was performed with the QIIME2 Emperor using the Bray-Curtis distance matrix to demonstrate the variation in microbial communities among the Kuflu cheese samples. Additionally, a Venn diagram was created to observe the differences of the identified bacterial and fungal genera in Kuflu cheese samples depending on the type of milk used in production. To establish the correlation between some parameters Pearson’s correlation coefficients were calculated using R software with the corrplot package (https://www.r-project.org).
The raw metagenome sequence data of bacterial and fungal profiles of Konya Kuflu cheese samples was added to the Sequence Read Archive (SRA) division of the National Centre for Biotechnology Information (NCBI) with the accession number of BioProject PRJNA993662 and BioProject PRJNA993704, respectively.
Results
The titratable acidity (TA), pH, moisture, salt in dry matter, and fat-in-dry matter of Kuflu cheese samples are shown in Table 1. The pH values ranged from 5.17 (MC10) to 6.69 (MC6) with an average value of 5.79. The TA values were found between 1.37 (MC6) and 2.81 g lactic acid/100 g cheese (MC10). The moisture content of the experimental Kuflu cheeses showed a high variability (in the range of 39.9%–56.7%). Turkish Standards Institute (TSI) reported that the salt content of mold-ripened cheeses could be at most 5% in dry matter, but it was determined that one of the experimental cheeses (MC8, 5.21%) had a salt content outside of this standard. In the MC5, MC2 and MC8 samples more fat in DM was detected than all other Kuflu cheeses, respectively 16.5%, 25%, and 27.5%.
In this study, we used two hyper-variable regions (V3 and V4) of the bacterial 16S rRNA and the number of clean reads obtained per sample ranged from 69,639 to 83,243 with an average length of 250 bases for Kuflu cheese samples. Also, ITS2 region was used for fungal microbiota and the number of clean reads obtained per samples ranged from 76,706 to 98,674 with an average length of 250 bases.
Fig. 2A indicates the relative abundances of bacteria in ten Kuflu cheeses at the phylum level. The predominant taxa at the phylum level were Firmicutes (63.89%–83.32%), Bacteriodetes (2.24%–21.44%), Proteobacteria (2.28%–17.80%), and Actinobacteria (which together accounted for more than 99.5% of the total relative bacterial abundance) in studied Kuflu cheese samples. Minor phylum which had low abundances with sequence frequencies of <0.5% included Verrucomicrobia, Fusobacteria, and Deinococcus-thermus, respectively. Although all cheese samples showed a similar bacterial diversity at the phylum level, the lowest Firmicutes abundances (63.89%, 64.18%, 68.78%) were observed in the MC7, MC9, and MC2 samples, respectively, whilst the highest Bacteriodetes abundances were in MC2 and MC10 samples (21.44% and 12.22%, respectively) and the highest Proteobacteria abundances were in MC7 and MC1 cheeses with 17.80% and 17.61%, respectively. Furthermore, the MC9 (15.22%), MC3 (14.06%), and MC7 (11.57%) cheeses contained greater proportions of Actinobacteria compared to the other samples while MC1 and MC10 samples had lower Actinobacteria phylum with 1.07% and 1.00% relative abundances, respectively.
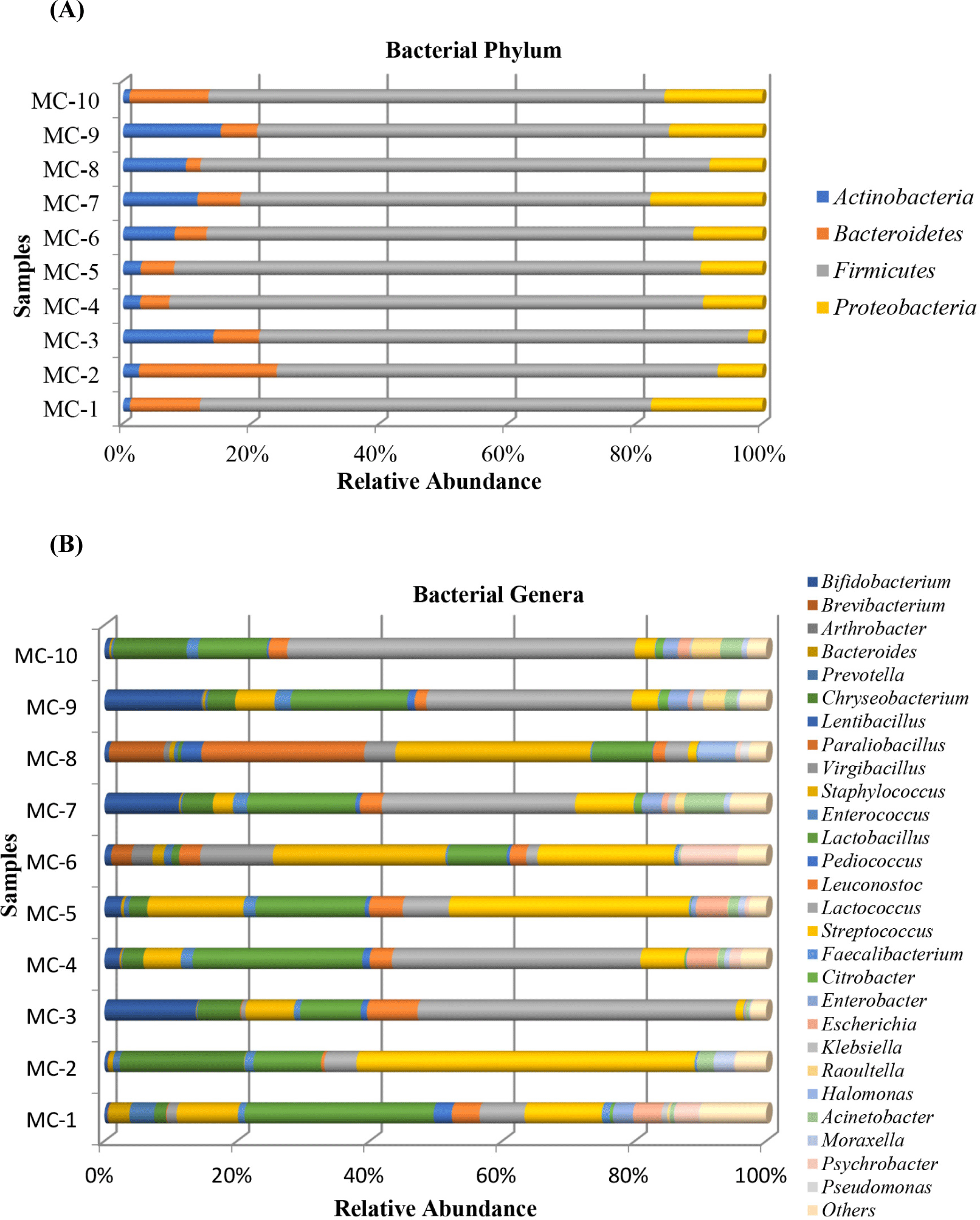
At the genus level, a total of 28 different genera were determined for 10 Kuflu cheese samples (Fig. 2B). Among these bacteria, Lactococcus genus dominated the microbiota of 5 cheese samples with relative abundances ranging from 29.18% (MC7) to 52.4% (MC10). Streptococcus genus showed dominance in 2 cheeses containing MC2 (51.03%) and MC5 (36.22%) samples, also Staphylococcus spp. were the highest genus in MC6 (26.03%) and MC8 (29.42%) cheeses. MC8 cheese showed a dissimilar bacterial proportion at the genus level, particularly with the highest abundance of Paraliobacillus (24.56%), Brevibacterium (8.23%), Halomonas (5.68%), and Virgibacillus (4.71%). As for the genus Virgibacillus, it was also present at a high relative abundance of 10.96% in MC6 samples. On the other hand, Chryseobacterium was relatively abundant in MC2 (18.75%) and MC10 (11.05%) cheeses among all studied samples. A large number of bacterial OTUs such as those affiliated with Bifidobacterium, Psychrobacter, Escherichia, Prevotella, and Acinetobacter genera was commonly detected in the Kuflu cheese samples.
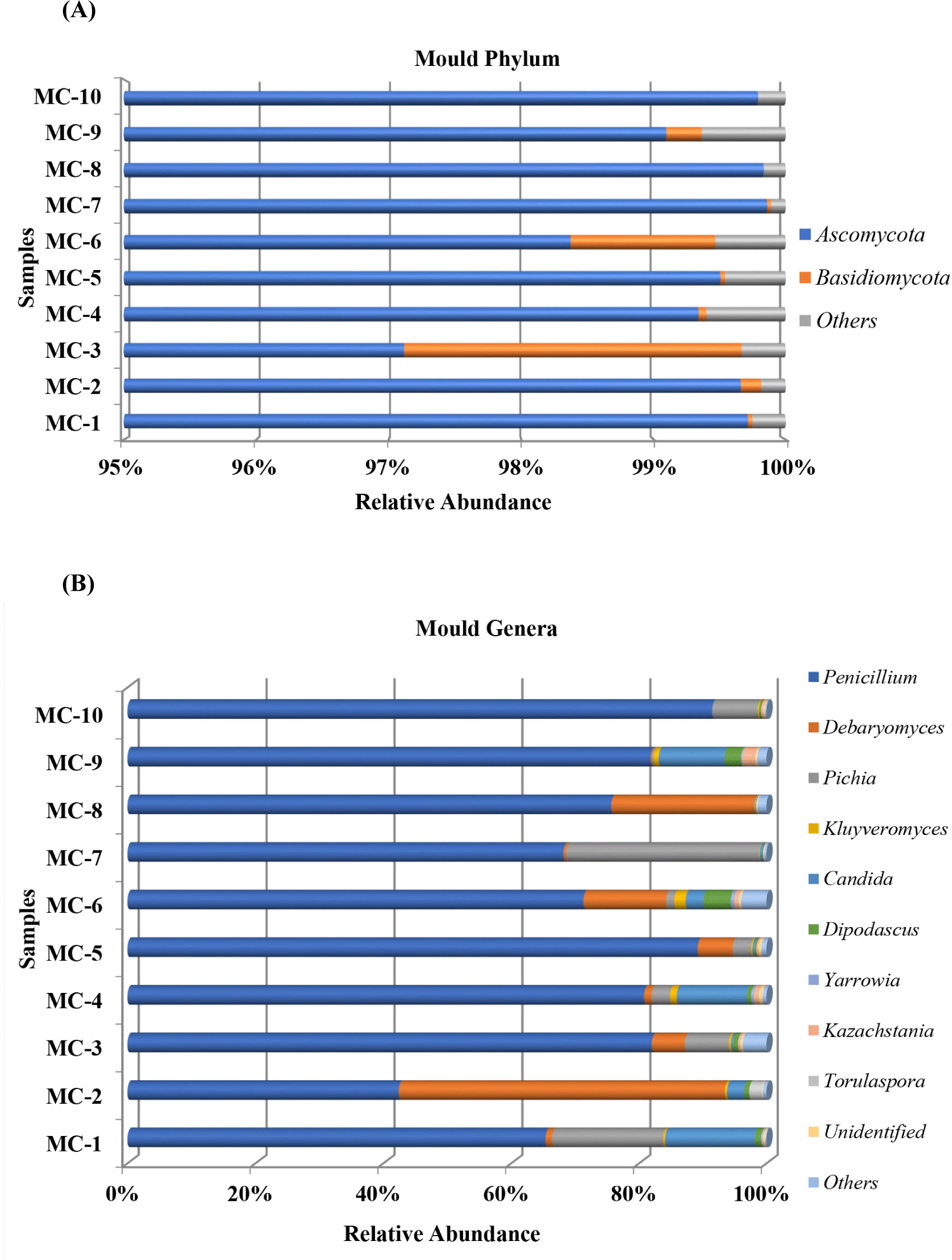
The taxonomic distribution of fungal biota at phylum level is shown in Fig. 3A. No important variation was reported for fungi diversity of ten Kuflu cheese samples and only two phyla were observed affiliated with Ascomycota and Basidiomycota. It was found that Ascomycota potently dominated the mycobiota of all cheese samples ranging from 97.11% (MC3) to 99.85% (MC7). Basidiomycota was detected relatively high in only two cheese samples (2.55% in MC3 and 1.09% in MC6), and for most cases, their prevalence was never >0.2%. Regarding fungi communities at the genus level, Penicillium dominated nine cheese samples with relative abundances ranging from 65.38% (MC1) to 91.46% (MC10) and in only one cheese sample (MC2) it was the second prevalent genus (42.42%) after the Debaryomyces genus (Fig. 3B). Pichia genus was more abundantly present in MC7 (30.28%) and MC1 (17.37%) cheeses than in all the other cheeses while its relative abundance showed differences ranging from 0.16% (MC8) to 7.17% (MC10) except for MC2 (not detected). As for Debaryomyces genus, MC2 cheese sample displayed higher abundances (51.03%). The genus Candida was the third prevalent genus in MC1 cheese with 14.08% relative abundance while it was the second in MC4 (10.99%) and MC9 (10.32%). Kluyveromyces (between 0.04% and 1.78% in all cheeses), Saccharomyces (between 0.03% and 0.57% in 7 cheeses), Fusarium (<0.1% in 6 cheeses) constituted the other fungi groups of Kuflu cheese samples, and Aspergillus genus, one of the important filamentous mold groups, were observed in only MC8 cheese (0.03%).
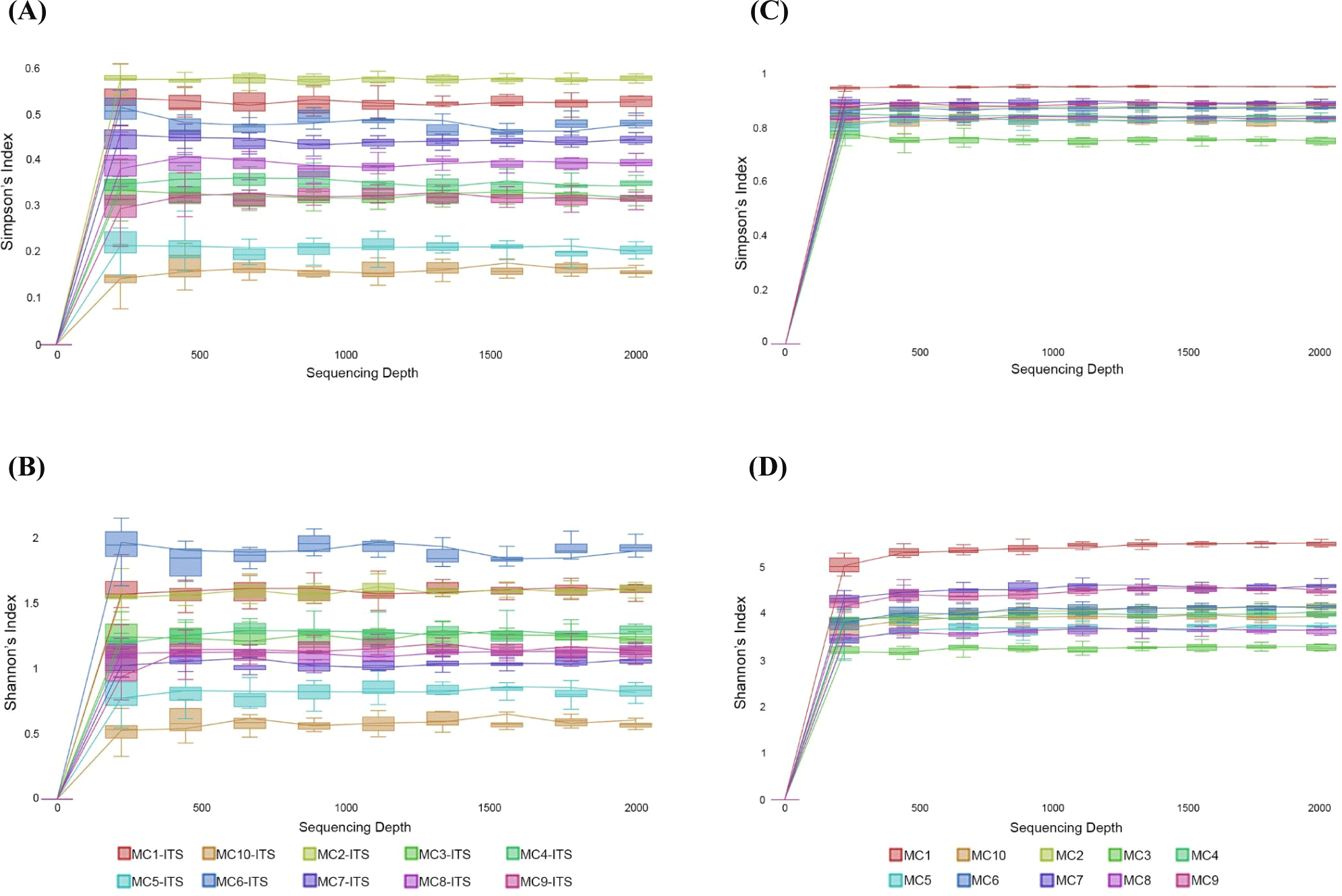
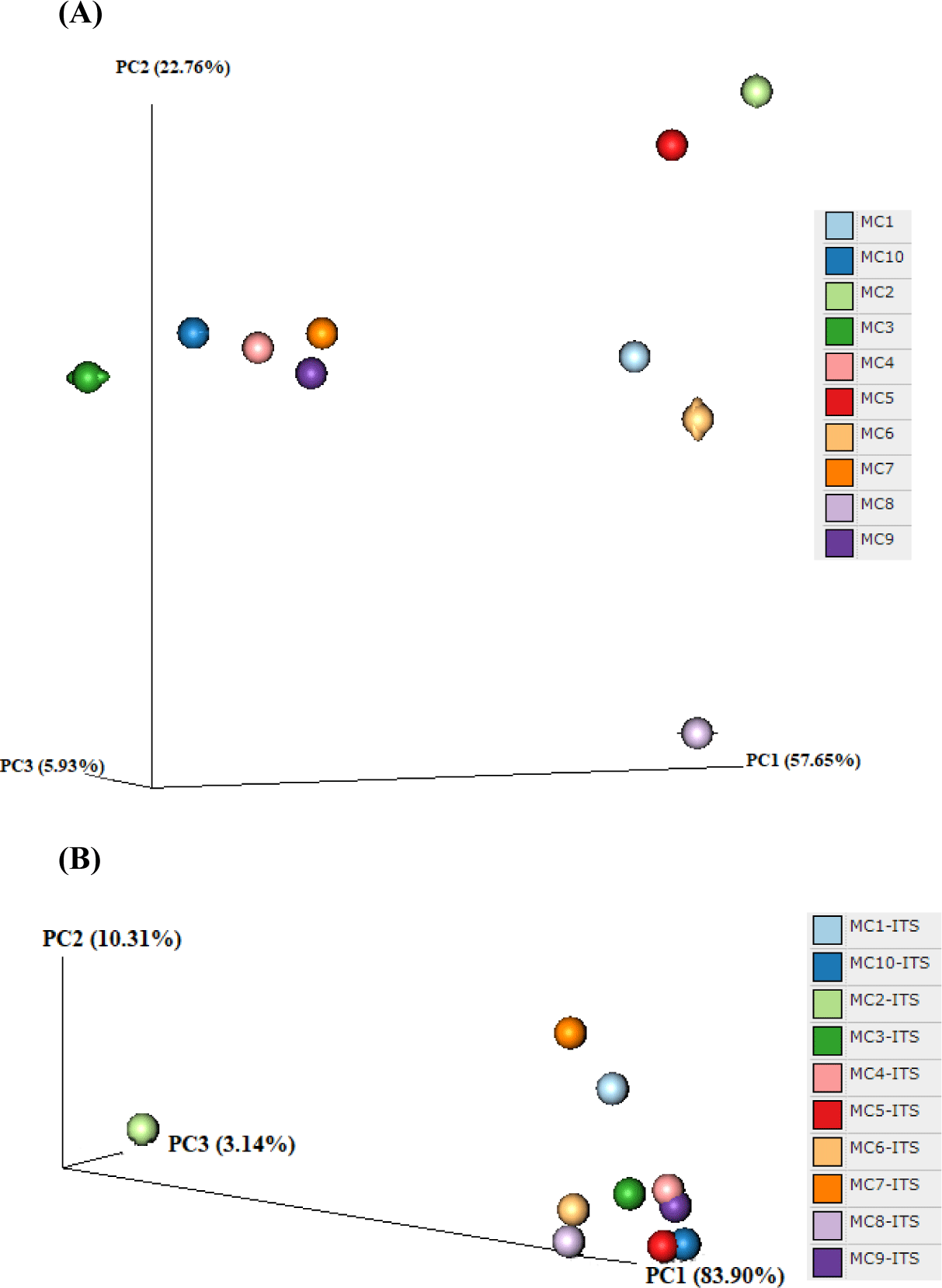
In the alpha diversity analysis, the Shannon and Simpson indices were calculated to estimate diversity and richness of the Kuflu cheese samples (Figs. 4A, B, C, and D). Alpha diversity indices reveal the structure of a bacterial community in terms of richness (number of OTU), evenness (distribution of abundances) or both. At this point, higher values of Shannon index indicate a greater genus diversity while lower Simpson indices show that a specific genera is dominant (Demirci et al., 2022). The greatest bacteria richness by Shannon index was detected in MC1 cheese sample (with a total 102 OTU-genera) while MC4 cheese presented the lowest Shannon index (with total 49 OTU-genera). As for Simpson index, MC3 cheese had the closest value to 0 (with 46.82% relative abundance for the most prevalent genus, which was Lactococcus) whereas MC1 cheese was located at the farthest point (with 11.11% relative abundance for the most predominant genera). Considering the fungal biota richness taken from Shannon index, MC6 cheese sample harbored a significantly greater richness and diversity with total of 35 different fungi OTUs. MC10 cheese had a very high dominance of one genus with a high proportion of sequences affiliated with Penicillium (91.46% of total OTU-genera) with the lowest Simpson index. To compare the similarities in microbiome compositions of each cheese sample, the beta diversity analysis was calculated and presented in the form of PCoA using Bray-Curtis dissimilarity indexes (Fig. 5). While the MC2 cheese sample differs from other cheeses in terms of bacterial community, MC3 cheese differs from the others in terms of fungal composition.
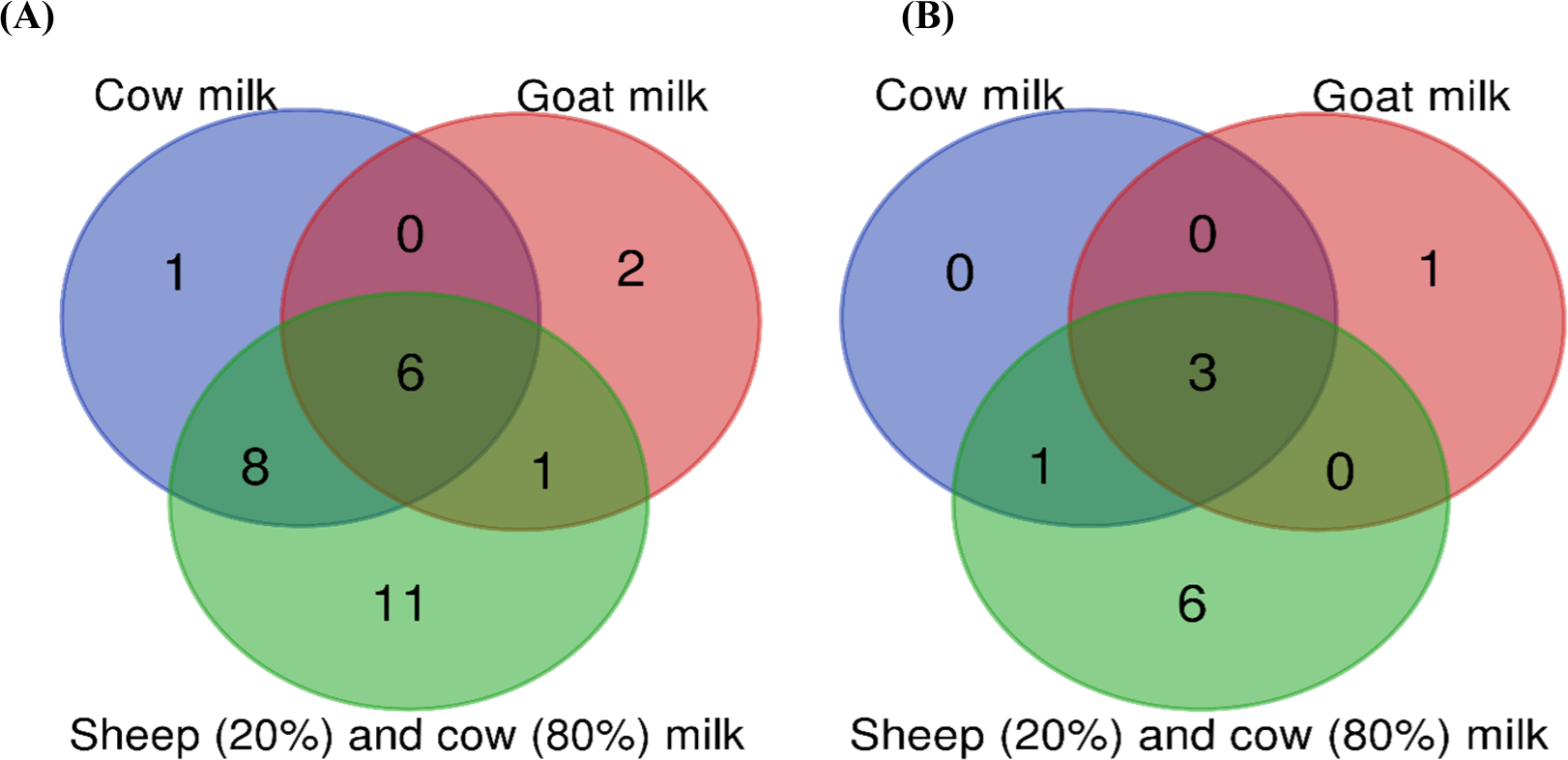
Venn diagrams were created to show the differences between the bacterial and fungal genera (>1% relative abundance) identified in the Kuflu cheeses produced from different types of milk (Fig. 6). A greater diversity in bacterial (Fig. 6A) and fungal (Fig. 6B) genera was observed in Kuflu cheeses produced using both sheep and cows according to the Venn chart. It was noted that the contribution of cow milk was the lowest in the diversity of microbial genera. Regarding bacterial genera, Lactobacillus, Lactococcus, Streptococcus, Enterococcus, Chryseobacterium, and Prevotella were common genera in all cheese varieties (Fig. 6A). As for fungal genera, Penicillium, Candida, and Debaryomyces genera were determined as joint groups (Fig. 6B). Kuflu cheeses, produced with a mixture of sheep-cow milk, contained 11 more bacterial genera and 6 more fungal genera. These differences reflect differences in the microbiota of the milk types used in the production (Butts et al., 2021).
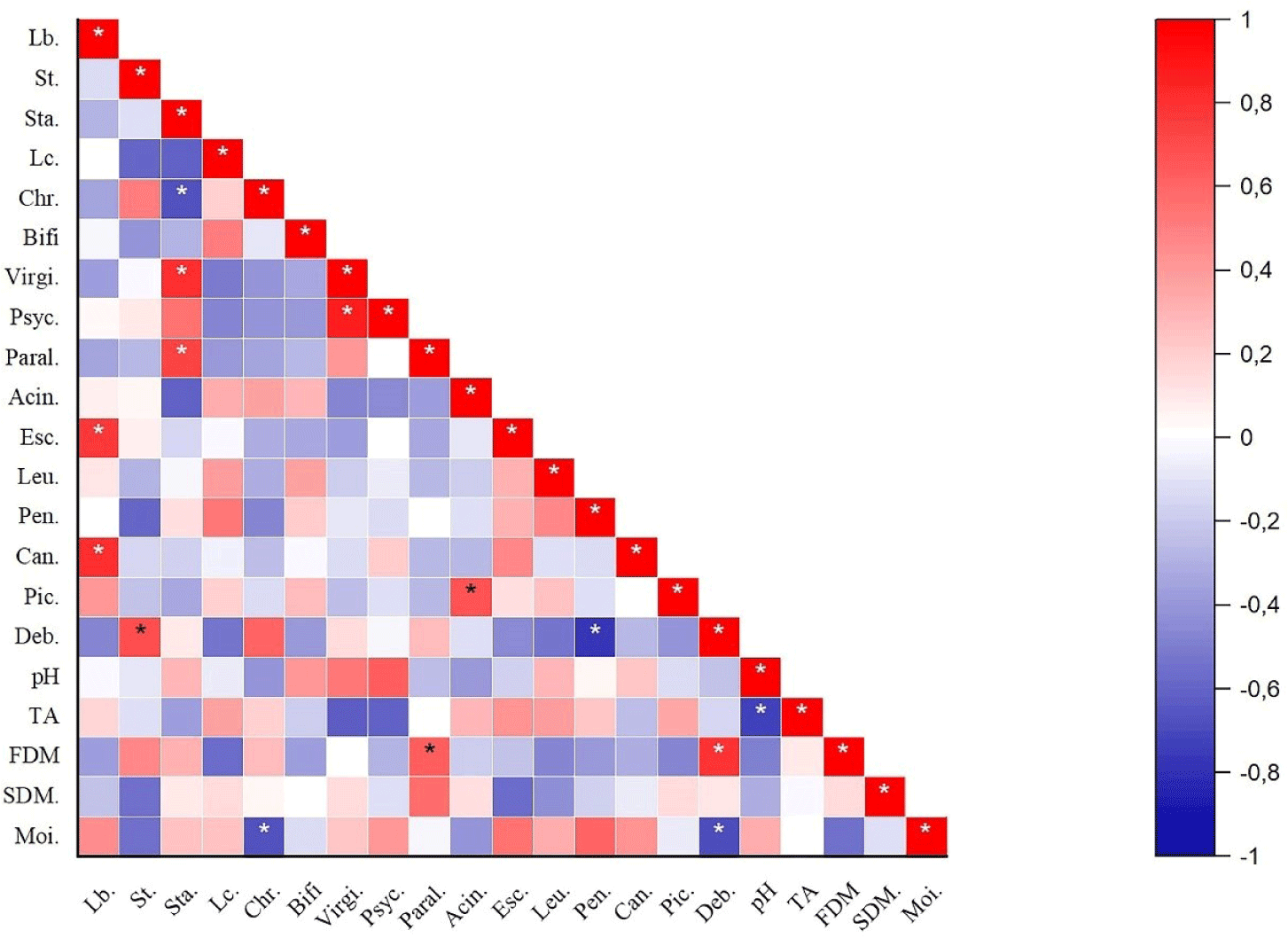
Correlation analysis between the relative abundance of dominant bacteria and fungal flora and pH, TA, and some chemical properties of Kuflu cheese samples was performed and significant correlations (p<0.05) were displayed in Fig. 7. The relative abundance of Lactobacillus genera was significantly positively correlated (p<0.05) with Escherichia and Candida spp. in Kuflu cheeses whilst Streptococcus genus was positively correlated with Debaryomyces. On the other hand, the abundance of Staphylococcus genus was significantly positively correlated (p<0.05) with Virgibacillus and Paraliobacillus abundances, whereas it was negatively correlated with Chryseobacterium. The relative abundance of Penicillium was also positively correlated (p<0.05) with Debaryomyces genera. Again, fat in dry matter was significantly positively correlated (p<0.05) with Paraliobacillus and Debaryomyces while moisture was negatively correlated with Chryseobacterium and Debaryomyces.
Discussion
Wide ranges in pH, TA, and chemical composition were determined in Kuflu cheeses. The pH values ranged from 5.17 (MC10) to 6.69 (MC6) with an average value of 5.79. The average pH value was found to be lower than that previously determined by Hayaloglu and Kirbag (2007) who studied on Kuflu cheese. Also, Hayaloglu et al. (2008) stated that the measured pH values were between 5.52 and 7.22, while the average value was 6.29. The differences in the regions where the cheeses are produced and in the raw materials and the differences arising from the manufacturer and production method may be the reason for this discrepancy. The average TA of Kuflu cheese samples (2.32 lactic acid/100 g) were higher than that reported by Hayaloglu et al. (2008) in line with pH results. The moisture content of the Kuflu cheeses showed a high variability (in the range of 39.9%–56.7%). Similar results were reported by Hayaloglu and Kirbag (2007) in Kuflu cheeses (37.65%–53.65%). TSI has been reported that the moisture content cannot be more than 45% in mold-ripened cheeses. From this point of view, it was determined that 9 out of 10 cheeses analyzed contained more moisture content than the value specified in the standard. Besides, TSI reported that the salt content of mold-ripened cheeses could be at most 5% in dry matter, but it was determined that MC8 cheese had a high salt content (5.21%) outside of this standard. In the MC5, MC2, and MC8 samples more fat in DM was detected than all other Kuflu cheeses, respectively 16.5%, 25%, and 27.5%, however, average fat in DM value (11.8%) was similar to the study conducted by Hayaloglu et al. (2008) who reported that the average fat in DM value was 12.18%. In fact, there can be serious discrepancies in the chemical compositions of such cheeses, which are traditionally produced under uncontrolled conditions, even in those manufactured in the same location. Indeed, Sengul et al. (2006) found very different results in the chemical composition of mold-ripened Civil cheeses and attributed this to the differences that may occur in the manufacturing procedures and ripening conditions of traditional raw milk cheeses.
In the present study, at phylum level, Firmicutes phylum showed dominance in all cheese samples with relative abundances ranging from 63.89% (MC7) to 83.32% (MC4) followed by Bacteriodetes with 2.24%–21.44% (MC8 and MC2, respectively) and Proteobacteria with 2.28%–17.61% (MC3 and MC1, respectively). Apart from these, the Actinobacteria constituted one of the important phyla in Kuflu cheese samples with 1.00%–14.06% (MC10 and MC3, respectively). In fact, several previous studies revealed that Firmicutes, Actinobacteria, Proteobacteria, and Bacteriodetes were predominating phyla in cheeses and also in ripened cheeses (Dugat-Bony et al., 2016), however, Unno et al. (2021) stated that bacterial biota of three types of surface mold-ripened cheeses (Brie de Meaux, Brie de Melun, and Coulommiers) were dominated by Firmicutes, followed by Proteobacteria, Actinobacteria, but not Bacteriodetes.
At the genus level, Lactococcus, Streptococcus, Lactobacillus, and Staphylococcus genera in general constituted the top four genera relative abundance with more than 50% -except for MC8- in Kuflu cheese samples, however, in some cheeses, Chryseobacterium (in MC10), Bifidobacterium (in MC3, MC7, and MC9), Paraliobacillus (in MC8), Virgibacillus (in MC6), Leuconostoc (MC3), and Brevibacterium (MC8) genera replaced one of them. Besides that, MC6 and MC8 cheese samples had a distinctive bacterial genera composition unlike the other cheeses because of the high prevalence of Virgibacillus (10.96%), Psycrobacter (8.65%), Paraliobacillus (3.17%) and Paraliobacillus (24.16%), Brevibacterium (8.23%), Halomonas (5.68%), Virgibacillus (4.71%), respectively. Regarding the taxonomic distribution of bacteria at genus level, Streptococcus spp. and Lactococcus spp., which are known as starter LAB, were the most dominant genera except for MC6 and MC8 cheeses which were dominated by staphylococci. This finding was not unexpected since Streptococcus spp. (mainly Streptococcus thermophilus) and Lactococcus spp. (mainly Lactococcus lactis) were previously identified among the most widespread and dominant genera in the untreated and ripened cheese microbiota with Leuconostoc spp., Lactiplantibacillus spp., and Enterococcus spp. by Montel et al. (2014). On the other hand, the genus Lactobacillus was found to be one of the most prevalent three genera in 9 out of the 10 Kuflu cheese samples with relative abundances changing between 9.07%–28.51% and it took the first and second places in MC1 (28.51%) and MC4 (25.59%) samples, respectively. Lactobacilli species -especially Lactobacillus delbrueckii subsp. bulgaricus- was used as a starter in blue-veined cheeses whilst facultatively heterofermentative species of the lactobacilli can be found in several mold-ripened and blue cheeses during ripening (Desmasures, 2014).
One of the most prevalent non-LAB detected in Kuflu cheeses was Staphylococcus genus which was found in MC6 (26.03%) and MC8 (29.42%) cheese samples with the highest presence amongst bacterial OTUs and again was present in MC5 and MC1 samples with 14.48% and 9.24% relative abundances, respectively. Many studies on fermented-food microbiota have reported that many coagulase-negative staphylococci are part of the indigenous microbiota which are originated from raw materials and food environment, and they are known to be food-related beneficial microbes. The presence of this genus at such high levels in mold ripened Kuflu cheeses may be due to the competitive characteristics attributed to many staphylococci owing to their capability to easily grow anaerobically at low water activity, in high concentration of NaCI, and at low temperatures. On the other hand, the genus Bifidobacterium was detected with increasing abundances in three cheese samples including MC7, MC3, and MC9 with 11.24%, 13.80%, and 14.65%, respectively. Additionally, this genus presented small but noticeable amounts in MC4 and MC5 cheese samples with 2.23% and 2.48%. Previously, this genus with probiotic potential was detected in cheeses made with similar production methods, such as Camembert, Brie, and Reblochon cheeses (Bondue et al., 2020). To the best of our knowledge, there is no report on the presence of Bifidobacterium in Turkish mold-ripened cheeses.
Surprisingly, the presence of the Paraliobacillus (24.56%) in MC8 cheese, which was marginally detected in other cheese samples except for MC6 (3.17%), was remarkably high. Paraliobacillus species have not been detected in any cheese before. In fact, the most striking feature of this group bacteria, which was detected at a high prevalence in only one Kuflu cheese sample, is that it needs to have high pH (pH 8) and NaCI (5%) concentrations for optimum growth (Chen et al., 2009). Indeed, the Kuflu cheese sample in which Paraliobacillus genus was detected very high (MC8), was the cheese sample with the highest NaCI in dry content among all samples (Table 1). Chryseobacterium genus was also detected in Kuflu cheese samples. This genera was reported in dairy products by previous researchers, for instance homemade yogurts (Demirci et al., 2022), Halloumi cheese (Kamilari et al., 2020), but it was not detected in Tulum or Kuflu cheeses in any study. Lastly, Chryseobacterium genus with a high relative abundance caused MC2 and MC10 cheeses to be distinguished from the other Kuflu cheese samples in terms of sensorial properties (Fig. 1).
With regard to fungi, at the phylum level, members of the Ascomycota heavily dominated all of the Kuflu cheese samples with relative abundances ranging from 97.11% to 99.85% corroborating the results already given by Dimov et al. (2021) who demonstrated that almost all fungi (99.9%) detected in Bulgarian green cheeses belonged to Ascomycota phylum. Nonetheless, in this present study, MC3 and MC6 samples differed from the other cheeses with a relatively high occurrence of Basidiomycota (2.55% in MC3 and 1.09% in MC6) which were mainly represented by the genera Panaeolus (1.3% in MC3) and Trichosporon (1.23% in MC3 and 0.95% in MC6). Although Panaeolus genus is an unexpected contaminant in cheese, Trichosporon has been detected in cheese before (Gelen and Ceylan, 2021).
Regarding the taxonomic distribution of fungi at genus level, members of the Penicillium genus were determined with increasing relative abundances in 9 out of the 10 Kuflu cheese samples ranging from 65.38% to 91.46% (in MC1 and MC10, respectively). These findings regarding the high abundances of Penicillium in mold-ripened Kuflu cheeses were confirmed by a recent study of Onmaz et al. (2021) in which the microbiota of mold-ripened Turkish cheeses. Indeed, this is an expected result since Penicillium spp. are known emblematic of blue-veined cheese varieties and they can be inoculated as a starter culture during cheese making process such as in Roquefort, Stilton, and Gorgonzola as well as appearing as the dominant filamentous mold of the environment in spontaneously molded cheeses (Desmasures, 2014; Montel et al., 2014). Differently, fungal community of MC2 cheese sample was dominated by Debaryomyces genus with 51.03% relative abundance followed by Penicillium genus (42.4%). Also, Debaryomyces genus was the second most prevalent fungi at genus level in MC8 and MC6 cheeses with 22.38% and 12.94% relative abundance, respectively. This may be attributed to the high fat content of MC2 and MC8 in dry matter, since Debaryomyces genus is known to be lipophilic (Song et al., 2022). It was previously reported that they were among the most important yeast genera for several cheese varieties, for example, Yildiz et al. (2021) showed that Debaryomyces genus –mostly Debaryomyces hansenii- was the most dominant in moldy Civil cheese followed by Pichia and Candida. On the other hand, many functional/probiotic properties are attributed to relevant yeast species such as antimicrobial killer toxin production, binding mycotoxin etc. (Esen and Çetin, 2021), therefore, the high rates of this genus in Kuflu cheeses are important in terms of the health benefits provided to the consumers of this cheese. Furthermore, two Kuflu cheese samples (MC7 and MC1) were characterized by an elevated representation of the Pichia genus with 30.28% and 17.37% relative abundances, respectively. Previous studies have reported that this yeast was one of the most prevalent genus in Bulgarian green cheeses (Dimov et al., 2021), traditional Turkish moldy Civil cheeses (Yildiz et al., 2021) and blue cheese varieties such as Roquefort, Cabrales, and Gorgonzola (Desmasures, 2014; Dimov et al., 2021; Yildiz et al., 2021).
As for Candida genus, it was found in all Kuflu cheese samples except for MC8 and MC10 and was amongst the most prevalent three yeast OTUs in MC1 (14.08%), MC4 (10.99%), and MC9 (10.32%). These are not surprising results, because this genus was reported to be one of the most common yeasts in several cheese varieties such as Tilsit, Reblochon, Surk, moldy Civil, Roquefort, Gorgonzola, Danablu, Bleu d’avuergne, Bleu de Bresse, and Valdeon cheeses (De Boer and Kuik, 1987). Surprisingly, one of the most remarkable findings of this study is the fact that Aspergillus genus, commonly associated with allergic bronchopulmonary aspergillosis producing potential, were not detected in any of the Kuflu cheese samples contrary to a study conducted by Onmaz et al. (2021) on mold-ripened Turkish cheese varieties. Besides, another finding of this study showed that Dipodascus genus was amongst the five most prevalent genera in half of the Kuflu cheese samples with relative abundances ranging from 0.29% to 2.58% and also Kluyveromyces genus did not constitute an important part of the yeast community of Kuflu cheese (0.1%–1.78%), which is different from the previous reports on mold-ripened cheeses (Desmasures, 2014; Yildiz et al., 2021). In fact, Kluyveromyces genus including especially Kluyveromyces marxianus and Kluyveromyces lactis, which were frequently detected in previous blue/mold-ripened cheese studies, were found in negligible amounts in the Kuflu cheese samples produced in Konya while Yarrowia genus was not detected at all (Dimov et al., 2021; Gkatzionis et al., 2014; Onmaz et al., 2021). The absence or low levels of these yeasts is a disadvantage for Kuflu cheeses since there are some strong data showing that these yeast species have probiotic properties and improve the aroma profile of cheeses as they are capable of producing high amounts of volatile compounds (Esen and Çetin, 2021). This may be due to the fact that these fungi are less salt-tolerant compared to D. hansenii and they cannot metabolize compounds, such as lactate, towards the end of ripening. It should also be noted that different bacterial and fungal genera that were not found in previous studies on Kuflu cheese, such as Virgibacillus, Paraliobacillus, and Bifidobacterium, were detected in the present study. Also, Dipodascus genus formerly Geotrichum, was detected in Kuflu cheeses with a percentage between 0.28% and 4.28% except for MC7 and MC8 samples. The lower abundance of this genus in Kuflu cheese as compared to other mold-ripened cheeses can be attributed to their salt sensitivity.
The greatest bacterial richness by Shannon index was detected in MC1 cheese sample with total 102 OTU-genera while MC4 cheese showed the lowest one with total 49 OTU-genera. Kuflu cheese samples containing an average of 62 bacterial OTU-genera had a better bacterial richness than Bulgarian green cheese according to the findings of Dimov et al. (2021). Regarding fungal richness by Shannon index, MC6 and MC3 cheeses had the highest OTUs at genus level (35 and 27 OTU-genera, respectively). MC10 cheese had a very high dominance of one genus with a high proportion of sequences belonging to Penicillium (91.46% of total OTU-genera) with the lowest Simpson index. Average fungal OTUs determined in this study are lower in comparison to the findings of Dimov et al. (2021) who studied on the Bulgarian green cheese batches.
As shown Fig. 7, the relative abundance of Lactobacillus genera was significantly positively correlated (p<0.05) with Candida in Kuflu cheeses. This is actually an unexpected result because there are studies showing that lactobacilli species have anti-candidal activities (Vazquez-Munoz and Dongari-Bagtzoglou, 2021). Although these studies are mostly related to the human opportunistic pathogen Candida albicans, in a study conducted on traditional white pickled and soft cheeses, very low abundance of lactobacilli were found in cheeses with a high abundance of Candida zeylanoides (Golić et al., 2013). Meanwhile, there are also studies that found a positive correlation between Candida and lactobacilli in parallel with this present study (Fujinami et al., 2021). Streptococcus genus was positively correlated with the relative abundance of Debaryomyces genus. Similarly, Stellato et al. (2015) observed that D. hansenii is also high in cheeses where S.thermophilus is high. On the other hand, Staphylococcus genus was significantly positively correlated (p<0.05) with Virgibacillus abundance whereas it was negatively correlated with Chryseobacterium. Positive interaction between staphylococci and Virgibacillus genus was previously observed in fish sauce (budu) produced using autochthonous Virgibacillus halodenitrificans PS21 and Staphylococcus simulans PMRS35 and in shrimp paste produced using Virgibacillus sp. SK37 and Staphylococcus nepalensis JS11 together (Kanjan et al., 2021; Yu et al., 2022). Again, a previous study revealed a positive correlation between Staphylococcus and Chryseobacterium in boiled milk samples in contrary to this current study (Joishy et al., 2019). This situation can be attributed to the possibility of dominance of different species in the genera mentioned in both studies. Also, the relative abundance of Penicillium was significantly positively correlated (p<0.05) with Debaryomyces genera in parallel with the findings of Stellato et al. (2015) who determined the positive correlation between Penicillium and Debaryomyces genera in cheese samples. In another study, the researchers have shown that D. hansenii, which is frequently found in blue cheeses, stimulates the growth of Penicillium roqueforti, that is, there is a positive interaction between them (Gkatzionis et al., 2014). The relative abundance of Paraliobacillus and Debaryomyces was significantly positively correlated (p<0.05) with fat in dry matter. This is no surprising result because especially Debaryomyces genera is known for having high lipolytic activity which may contribute to the flavor and texture development of fermented products (Song et al., 2022). Moisture was negatively correlated with the relative abundance of Debaryomyces and Chryseobacterium. Debaryomyces species, especially D. hansenii, is particularly tolerant of high salt or high sugar environments, therefore, its negative correlation with moisture content is not surprising.
Conclusion
As a result of the study, Firmicutes was found as the most dominant bacterial phyla, while Lactococcus, Streptococcus, and Staphylooccus were detected as the most dominant bacterial genera, respectively. The genus Virgibacillus, which had not been detected in mold-ripened cheeses before, was detected with high relative abundances in some Kuflu cheeses. As for the fungal composition, Ascomycota was very dominant at the phylum level as expected, and the genus Penicillium was found to be the most prevalent in most of the cheeses at genus level. No OTU belonging to Aspergillus was detected in Kuflu cheeses. At the same time, none of the important food pathogens were detected in these cheeses. This study enlightens the researchers and producers interested in mold-ripened cheeses and Kuflu cheese, and it is also important to improve the quality of this uncontrolled moldy cheese variety. Overall, this work may be an initial study to exhibit the benefits and risks of Kuflu cheeses in the context of microbial composition using metagenomic data, and it has also shown that Kuflu cheeses can assist in the isolation and identification of the first detected strains in Kuflu cheeses such as Bifidobacterium and Lactobacillus species using culture-dependent methods.