Introduction
Methicillin-resistant Staphylococcus aureus (MRSA) has become one of the most significant pathogens worldwide (Goudarzi et al., 2017). MRSA exhibits resistance to penicillin and other β-lactam antibiotics due to the presence of the mecA gene encoding the penicillin-binding protein 2a (PBP2a) (Chambers, 1997). MRSA presents a major health concern in humans (Tacconelli, 2009; Tiemersma et al., 2004).
The presence of MRSA strains in food-producing animals has led to concerns regarding foodborne contamination (Weese, 2010); MRSA has been identified in retail meat including beef, veal, lamb, pork, and poultry worldwide (Dhup et al., 2015). Particular focus has been on pork products, as livestock-associated MRSA (LA-MRSA) is known to colonize pigs (Peeters et al., 2015).
Food products of animal origin play an important role in antimicrobial-resistance dissemination (Muloi et al., 2018). Marshall and Levy (2011) reported that the use of antibiotics in food-production animals, especially for non-therapeutic use, is linked to their resistance in people living on or near farms, and even in the general population via the food chain. Transmission of MRSA from food animals to humans was first realized in case of swine, since pig farmers and family members were found to be infected with MRSA sequence type (ST) 398 (Voss et al., 2005). Since then, studies worldwide have documented the presence of MRSA in various food-producing animals and people in frequent contact with these animals (Fitzgerald, 2012). In Korea, MRSA has been reported in nonclinical sources, including pigs (Lim et al., 2012), milk (Song et al., 2016), and dogs (Kwon et al., 2006), as well as clinical isolates (Joo et al., 2017; Kim et al., 2018; Moon et al., 2015). However, little is known about the presence of antimicrobial-resistant S. aureus and MRSA from retail meats in Korea. Therefore, aim of this study is to investigate the prevalence of antimicrobial-resistant S. aureus and MRSA strains from retail meats (beef, pork, and chicken) in Korea and their molecular characteristics.
Materials and Methods
A total of 4,264 meat samples were purchased from retail markets and supermarkets in Korea from 2013 to 2018. Among the 2,049 domestic meat products, beef, pork, and chicken accounted for 719, 671, and 659 samples, respectively. The total number of imported meat products was 2,215, which included 868 beef (Australia 505, USA: 297, New Zealand 38, Mexico 15, and Canada 13), 789 pork (USA 171, Spain 169, Mexico 120, Germany 102, Canada 57, Chile 47, Belgium 29, Netherlands 21, Austria 20, Denmark 17, France 15, Ireland 7, Finland 5, Poland 5, Hungary 3, and Australia 1), and 558 chicken samples (Brazil 422, USA 100, Denmark 26, Thailand 8, and Netherlands 2).
For the isolation of S. aureus, 25 g of the meat sample and 225 mL of tryptic soy broth (Oxoid, Cambridge, UK) supplemented with 10% NaCl were mixed and incubated at 37°C for 18 to 24 h. One loop of the enrichment broth was then streaked on Baird-Parker RPF (Oxoid) medium and cultured for 24 to 48 h. A typical colony—black in color on turbid white circles—was selected and the species was identified on tryptic soy agar (TSA) (Oxoid) using VITEK MS system (bio-Meriuex, Marcy I’Etoile, France).
Antimicrobial susceptibility tests for S. aureus (n=777) were carried out using the minimum inhibitory concentration (MIC) method. For the MIC test, a custom-made EUST plate (Thermo Trek Diagnostics, OH, USA) was used. The inoculum of S. aureus was prepared from overnight (18 h) growth on TSA and diluted to a 0.5 McFarland turbidity concentration with 0.45% saline. This suspension was further diluted with cation-adjusted Mueller-Hinton broth with N-tris(hydroxymethul) methyl-2-aminoethanesulfonic acid (TES) (CAMHBT; Remel, Lenexa, KS, USA) to obtain a final concentration of 5×104 CFU/well. This dilute was dispensed into each well of EUST plate and MICs were determined after incubation at 37°C for 18 h. The antibiotics used in the MIC test and the test ranges are given in Table 1. Susceptibility results in the form of MICs were interpreted by referring to the Clinical and Laboratory Standards Institute (CLSI) guidelines and European Committee on Antimicrobial Susceptibility Testing (EUCAST) guidelines (Table 1). For confirmation of MRSA, only strains showing resistance to cefoxitin were selected.
Cefoxitin-resistant S. aureus isolates (n=64) were subjected to DNA extraction followed by amplification of mecA gene by PCR using the primers mecA-F (5'- AAAATCGATGGTAAAGGTTGGC-3') and mecA-R (5'-AGTTCTGCAGTACCGGATT TGC-3') (Datta et al., 2011). The reaction mixture (20 μL) consisted of 10 pmol of each primer, Taq DNA polymerase (1 U), dNTP (dATP, dCTP, dGTP, dTTP) (250 μM each), reaction buffer with 1.5 mM MgCl2, and 2 μL template DNA. The amplification was performed by denaturation at 94°C for 30 s, annealing at 58°C for 1 min, extension at 72°C for 1 min, and a final extension at 72°C for 10 min. The amplified product was a 533 bp sequence, which was resolved by electrophoresis on a 1.5% agarose gel and observed under UV light. All strains positive for the mecA gene were designated MRSA.
Multilocus sequence typing (MLST) was performed as described previously (Enright et al., 2000). The housekeeping gene primers used in the MLST analysis are given in Table 2. Alleles and ST were assigned by submitting the DNA sequences to the Staphylococcus MLST database (http://saureus.mlst.net). Amplification of the polymorphic region of the Staphylococcus protein A gene (spa typing) was performed according to the protocol suggested previously (Strommenger et al., 2006). Primers spa-1113f (5'-TAAAGACGATCCTTCGGTGAGC-3') and spa-1514r (5'-CAGCAGTAGTGCCGTTTGCTT-3') were used for amplification. The thermal cycling reactions consisted of an initial denaturation at 80°C for 5 min, followed by 35 cycles of denaturation at 94°C for 45 s, annealing at 60°C for 45 s, and extension at 72°C for 90 s. The sequence of the PCR product was analyzed by a ABI3730xl DNA analyzer (Applied Biosystems, CA, USA). All sequencing reactions were performed with 25 cycles of denaturation at 96°C for 10 s, and extension at 60°C for 4 min. The isolates were then assigned to the specific spa types according to the guidelines described on the Ridom SpaServer database (http://www.spaserver.ridom.de).
| Housekeeping gene | Size (bp) | Primer sequence | Reference |
|---|---|---|---|
| arc | 456 | F 5'- TTGATTCACCAGCGCGTATTGTC -3' R 5'- AGGTATCTGCTTCAATCAGCG -3' |
Enright et al. (2000) |
| aroE | 456 | F 5'- ATCGGAAATCCTATTTCACATTC -3' R 5'- GGTGTTGTATTAATAACGATATC -3' |
|
| glpF | 465 | F 5'- CTAGGAACTGCAATCTTAATCC -3' R 5'- TGGTAAAATCGCATGTCCAATTC -3' |
|
| gmk | 429 | F 5'- ATCGTTTTATCGGGACCATC -3' R 5'- TCATTAACTACAACGTAATCGTA -3' |
|
| pta | 474 | F 5'- GTTAAAATCGTATTACCTGAAGG -3' R 5'- GACCCTTTTGTTGAAAAGCTTAA -3' |
|
| tpi | 402 | F 5'- TCGTTCATTCTGAACGTCGTGAA -3' R 5'- TTTGCACCTTCTAACAATTGTAC -3' |
|
| yqiL | 516 | F 5'- CAGCATACAGGACACCTATTGGC -3' R 5'- CGTTGAGGAATCGATACTGGAAC -3' |
The 18 non-ST398 MRSA isolates were subjected to pulsed-field gel electrophoresis (PFGE) analysis. Plugs digested with SmaI were run on 1.2% SeaKem Gold agarose gel (Lonza, Basel, Switzerland) with 0.5x Tris-Borate EDTA (TBE) buffer (Bioneer, Korea) on the CHEF Mapper PFGE system (Bio-Rad, CA, USA) for 18 h at 14°C, with an initial and final switch time of 5.16 s and 40.17 s, respectively. Salmonella enterica serovar Braenderup digested with XbaI was used as the molecular reference marker. Gels were stained with SYBR Gold solution (Invitrogen, CA, USA), and viewed and recorded under a UV transilluminator. The images were analyzed using the BioNumerics software (Applied Maths, TX, USA).
The statistical significance of differences between proportions was evaluated with Chi-square (χ2) tests using Epitools (Sergeant, 2018).
Results and Discussion
The prevalence of S. aureus and MRSA in retail meat samples is shown in Table 3. A total of 777 S. aureus strains were isolated from 4,264 raw retail meat samples. The contamination with S. aureus was the most in chicken meat among the domestic samples (33.2%) (p<0.05). The prevalence of S. aureus contamination in chicken carcasses was also highest compared with other food-producing animal carcasses in the previous report and was lower than that in our study (NIFDS et al., 2019). This suggests that the cross contamination may occur more frequently during the manufacturing and distribution processes (such as slaughter, cutting, packaging, or transportation) in the chicken meat supply chain, compared with other meat products. On the other hand, of the imported samples, S. aureus was the most detected in pork (20.9%) (p<0.05). The prevalence of S. aureus in the domestic meat samples was significantly higher than that in the imported samples (p=0.0008). Previous studies have shown that the isolation rate of S. aureus in retail meat was 35.0% (647/1,850) in China (Wu et al., 2018), 27.9% (65/289) in USA (Ge et al., 2017), and 21.8% (89/408) in India (Zehra et al., 2019). In Korea, the isolation rates of 35.4% (74/209) (Cho et al., 2014) and 11.0% (218/1,984) (Heo et al., 2008) have been reported. Most of the above studies observed isolation rates higher than that of our results, but they also showed varying isolation rates depending on the type of meat product and processing technique. In case of imported meat products, it is thought that the possibility of survival of the strains is lower due to the frequent freezing during distribution.
1) Difference between the S. aureus positive proportions of domestic and imported meat by Chi-squared test.
Among the 777 S. aureus strains, 64 cefoxitin-resistant strains were tested for the mecA gene, and 29 strains (0.7%) were identified as MRSA (Table 3). Year-wise, 6 strains were identified in 2013, 2 were identified in 2014, 5 were identified in 2015, 2 were identified in 2016, 9 were identified in 2017, and 5 were identified in 2018. Our study indicates that there were no significant differences in the year-wise prevalence of MRSA (p>0.05) (data not shown). According to the type of meat products, MRSA derived from pork was the most common at 19 strains, and the remainder was 6 strains from chicken and 4 from beef. No significant difference in the prevalence of MRSA strains was present between domestic and imported meat samples (p=0.8333). The prevalence of MRSA in this study was similar to that reported in Japan (0.9%, 1/115) (Baba et al., 2010), Malaysia (1.4%, 5/360) (Neela et al., 2009), and Korea (0.5%, 13/2,810) (Kim et al., 2015). However, studies showing higher prevalence have been reported in the Netherlands (11.9%, 264/2,217) (de Boer et al., 2009) and Hong Kong (10.6%, 126/1,190) (Boost et al., 2013). According to Ge et al. (2017), the prevalence of MRSA in retail meat may vary widely depending on the geographical location. Even at such low levels, MRSA may be present in meat, and in conditions where the treatment of contaminated food is inadequate, it can be transmitted to humans where it can colonize the nasal passages, skin, and the gastrointestinal tract (de Boer et al., 2009).
This study has a limitation regarding the confirmation of MRSA isolates. Because mecC-positive strains were not determined, the prevalence of MRSA in our study could be underestimated. mecC (formerly mecALGA251) has been identified as a mecA homolog with ~69% similarity to the mecA gene at the DNA level (Peterson et al., 2014). In future studies, it will be necessary to monitor the presence of mecA and mecC genes for the identification of MRSA because of its divergence from mecA (Ariza-Miguel et al., 2014).
777 S. aureus strains were tested for resistance to 17 antibiotics (Table 4). Resistance to penicillin was observed in 441 of the 777 strains (56.8%), which was the highest among all the antibiotics. The resistance rates against tetracycline and erythromycin were 27.4% and 23.9%, respectively. On the other hand, all strains showed susceptibility to linezolid, rifampin, and vancomycin.
Overall, in the case of domestic samples, the resistance rates of S. aureus from beef were lower than that of pork and chicken. However, in case of penicillin and fusidate, the resistance rate for the strain derived from beef and pork was higher than that isolated from chicken (p<0.05). The national sales data for veterinary antibiotics have indicated that the sales of antibiotics for swine and poultry breeding (492 tons and 158 tons, respectively) were more than that for cattle (92 tons) in 2018 (NIFDS et al., 2019). This may explain why resistance in S. aureus from pork and chicken meat was higher than that from beef. Moreover, the types of antibiotics used in each animal breeding were different (NIFDS et al., 2019). This may explain why the antimicrobial resistance profiles of S. aureus were different in each meat category. The resistance rates of S. aureus from the imported samples were not significantly different among meat categories except for penicillin, tetracycline, and clindamycin.
The resistance rate of S. aureus to gentamicin, kanamycin, penicillin, ciprofloxacin, and chloramphenicol in domestic sample isolates was significantly higher than those in imported isolates (p<0.05). Conversely, the resistances of S. aureus from imported samples to sulfamethoxazole and mupirocin were higher than those from domestic samples (p<0.05). No significant difference in the resistance of S. aureus strains against other antibiotics was present between domestic and imported samples.
Antimicrobial susceptibility profiles revealed that the 29 MRSA strains were resistant to penicillin and cefoxitin (Table 5). Additionally, the strains exhibited relatively high resistance rates against tetracycline (62.1%), clindamycin (55.2%), and erythromycin (55.2%). Similar to previous reports, ST398 was observed to be resistant to tetracycline in this study (Fluit, 2012; Vanderhaeghen et al., 2010). However, all 29 strains were susceptible to rifampin, vancomycin, linezolid, and mupirocin.
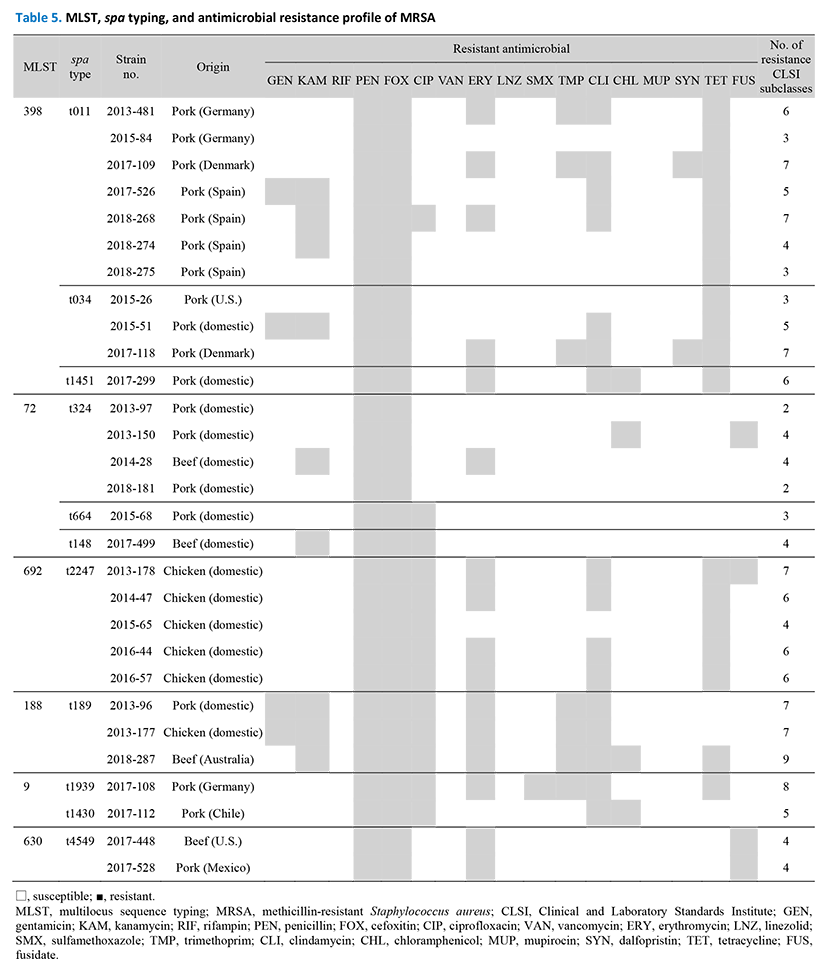
MLST, multilocus sequence typing; MRSA, methicillin-resistant Staphylococcus aureus; CLSI, Clinical and Laboratory Standards Institute; GEN, gentamicin; KAM, kanamycin; RIF, rifampin; PEN, penicillin; FOX, cefoxitin; CIP, ciprofloxacin; VAN, vancomycin; ERY, erythromycin; LNZ, linezolid; SMX, sulfamethoxazole; TMP, trimethoprim; CLI, clindamycin; CHL, chloramphenicol; MUP, mupirocin; SYN, dalfopristin; TET, tetracycline; FUS, fusidate.
Based on CLSI subclasses criteria, 27 out of 29 MRSA strains (93.1%) were multidrug-resistant MRSA strains (Table 5). In several studies (Abdalrahman et al., 2015; Fox et al., 2017; Li et al., 2017; Normanno et al., 2015), a large proportion of MRSA strains isolated from meat were also revealed to be multidrug-resistant. Ge et al. (2017) reported that multidrug-resistance (MDR) rates in MRSA (37.2%, 29/78) were higher than those of methicillin-susceptible Staphylococcus aureus (MSSA) (8.2%, 78/954). This is consistent with our result showing that MDR in MRSA was significantly higher than that in MSSA (p<0.01) (data not shown). Farm animals could be an important ecological niche for the emergence of multidrug-resistant S. aureus, since the massive use of antibiotics for treatment, prevention of diseases, or growth promotion provides the necessary evolutionary constraints (Yan et al., 2014). In addition, with regard to public infections, a multidrug-resistant pathogen is an emerging concern among all kinds of meat (Petternel et al., 2014).
The results of PFGE with SmaI for the 18 strains, except ST398, are shown in Fig. 1. It is known that ST398 are nontypeable using PFGE with SmaI (Huijsdens et al., 2006). The results obtained from the 18 strains showed 15 pulsotypes based on 100% similarity. Among them, there were three pairs (2016-44 and 2016-57, 2017-448 and 2017-528, and 2013-97 and 2013-150) which displayed 100% homology, and whose MLST and spa type also coincided (ST692-t2247, ST630-t4549, and ST72-t324). Two strains of ST692 (2016-44 and 2016-57) with 100% similarity were isolated from chicken meat products of different companies. Two strains of ST72 with the same pulsotype (2013-97 and 2013-150) were isolated from pork meat of different companies. Two strains of ST630 (2017-448 and 2017-528) with the same pulsotype were isolated from US beef and Mexican pork, respectively. Therefore, the contaminating origin of these MRSA pairs with the same pulsotype was impossible to be tracked.

The results of MLST and spa typing analysis for the 29 MRSA are shown in Table 5. The majority (37.9%) belonged to ST398, including 7 strains of t011, 3 strains of t034, and 1 strain of t1451. All ST398 strains were from imported pork, except for two, which were from domestic pork. The antibiotic resistance profile for ST398 showed the same pattern as that observed in the two strains derived from Danish pork in 2017. In addition, the resistance profiles of the strain derived from Spanish pork and that from domestic pork coincided with each other. ST398 is known to have been isolated from pig nasal passages in many European countries, including Germany, Austria (Witte et al., 2007) and Portugal (Conceição et al., 2017). In the Netherlands, ST398 was detected not only in pigs, but also in humans who were in regular contact with pigs (Voss et al., 2005). The clonal lineage of ST398 was also detected in Korean pigs (Eom et al., 2019; Lim et al., 2012). Our study suggests that ST398 strains from retail pork meat come from porcine origins, not from humans.
Out of the six strains analyzed by ST72, four strains were determined as t324; the other two were determined as t664 and t148. All strains identified as t324 were derived from domestic pork or beef, and the antimicrobial resistance patterns were inconsistent, except for those resistant to penicillin and cefoxitin. However, all strains of ST72 showed susceptibility to tetracycline. Several studies (Kim et al., 2020; Lim et al., 2012; Park et al., 2007) also reported that the ST72 type isolates were susceptible to tetracycline. ST72 is the most reported type in Korea, and is widely prevalent in clinical environment, livestock products, and soil (Ko et al., 2011; Lim et al., 2010). Other studies have referred to ST72 as a major community-associated MRSA in Korea (Kim et al., 2007; Ko et al., 2008; Park et al., 2015). In this study, since 2013, it has been consistently separated only from domestic meats.
All five strains of ST692 were identified as t2247, and originated from domestic chicken. Three of the strains showed the same pattern of antibiotic resistance. Moon et al. (2015) reported that ST692 from chicken carcasses was the LA-MRSA type in Korea. MRSA of the ST692 type has also been isolated from domestic chicken previously (Lim et al., 2010). Based on these findings, ST692 may be chicken-specific lineage of LA-MRSA in Korea.
Three strains of ST188 were revealed to be t189, and two of these strains from domestic samples in 2013 had the same antibiotic resistance pattern. The other strain was isolated from Australian beef in 2018. ST188 is the most prevalent type from clinical case in Asia and the Pacific region; it has been detected in Hong Kong (Monecke et al., 2011), Malaysia (Ghaznavi-Rad et al., 2010), and Taiwan (Chen et al., 2012). ST188 has been reported as the community-associated MRSA (CA-MRSA) in Australia (Nimmo and Coombs, 2008). Although rarely reported as the LA-MRSA, ST188 has been reported to be a major clonal type of human and livestock infections in Shanghai, China (Wang et al., 2018).
Two strains of ST630 also showed the same antibiotic resistance pattern, despite the fact that they were isolated from different meat sources and were from countries. Two strains revealed as ST9 showed different spa types (t1939 and t1430), as well as different resistance patterns.
spa typing is a DNA sequencing method for the mutation of the X-region of the Protein-A gene, and is particularly useful for distinguishing strains that cannot be distinguished using PFGE (de Boer et al., 2009). In addition, molecular typing using MLST and spa typing is advantageous for S. aureus strain typing due to the reproducibility of results, ease of use, and exchangeability (Strommenger et al., 2008). As shown in the present study, spa typing results are highly consistent with those of MLST (O’Hara et al., 2016).
To summarize, we performed prevalence studies for S. aureus and genetic characterization of MRSA in retail meat from Korea. The identification of MRSA in the final food consumption stage, which has been isolated at a low rate, is indicative of potential risks, and its presence or absence must be closely monitored at the national level. In addition, future studies may be directed towards monitoring the distribution and transmission of healthcare-associated (HA) MRSA, CA-MRSA, and LA-MRSA in farms, slaughterhouses, and retail foods.













