Introduction
Staphylococcus aureus is a primary cause of human infections and one of the most important foodborne pathogens (Hennekinne et al., 2012; Turner et al., 2019). S. aureus has been implicated in a number of infectious diseases such as minor skin and soft tissue infections, toxic shock syndrome, and septicemia (Turner et al., 2019). Coupled with the virulence, S. aureus can also develop antimicrobial resistance to methicillin (methicillin-resistant S. aureus; MRSA), vancomycin (vancomycin-intermediate or -resistant S. aureus; VISA/VRSA), and daptomycin (Turner et al., 2019). In addition to the infections in humans, a recent increase in the occurrence of livestock-associated (LA)-MRSA has been observed in various livestock animals, including the prevalence of sequence type (ST) 398 LA-MRSA in pigs worldwide (Price et al., 2012). The LA-MRSA strains tend to develop multidrug resistance (MDR) to several classes of antimicrobials, particularly tetracycline (Larsen et al., 2016; Price et al., 2012). Although there has been less focus on methicillin-susceptible S. aureus (MSSA) than MRSA, an increase in MSSA infections has been reported both in humans and animals (Carfora et al., 2016). Because MRSA and MSSA in livestock can be transmitted to farm workers and foods of animal origin and thus to CA settings, several studies were performed to monitor the prevalence of MRSA and MSSA in chicken, pork, and beef meat samples (Moon et al., 2015; Osman et al., 2016; Pauly et al., 2019).
Although the prevalence of MRSA and MSSA in swine farms and pork has been studied extensively (Moon et al., 2015), information is lacking on the occurrence and genetic profiles of MRSA and MSSA in the beef production system (retail markets, slaughterhouses, and beef cattle farms). Previous studies on bovine-associated MRSA or MSSA have mostly focused on bovine mastitis in the dairy industry (Song et al., 2016; Vanderhaeghen et al., 2010). Song et al. (2016) characterized profiles of antimicrobial resistance and staphylococcal enterotoxin (SE) genes in MRSA strains isolated from mastitic milk samples in Korea. Although several previous studies also identified MRSA in major livestock animals and meat samples in Korea (Lim et al., 2010; Moon et al., 2015), to the best of our knowledge, this is the first study in Korea that performed a nationwide screening of both MRSA and MSSA in the beef production system including beef cattle, beef meat, farmers or workers, and facilities.
In this study, we investigated the prevalence and genotypes of MRSA and MSSA throughout the beef production system in Korea, encompassing beef cattle farms, retail markets, and slaughterhouses. In addition, we examined genotypic and phenotypic factors associated with antimicrobial resistance and analyzed virulence characteristics of the MRSA and MSSA isolates.
Materials and Methods
A total of 1,148 swabs and 137 meat samples were obtained from the following: 20 beef cattle farms, 7 slaughterhouses, and 20 retail markets from eight different provinces of Korea during 2018. Samples from beef cattle farm were obtained from healthy cattle (n=169), the cattle farm environment (n=60), and farmers (n=21); retail market samples included fresh beef meat (n=137), the environments within retail facility (n=13), and retail market workers (n=7); slaughterhouse samples were collected from cattle carcasses (n=264), the facility environments (n=21), and workers (n=9). All samples were transported to the laboratory within 24 h of sampling for isolation of S. aureus.
The swab samples were inoculated into 5 mL of fresh tryptic soy broth (TSB; Difco Laboratories, Detroit, MI, USA) containing 10% NaCl (NaCl-TSB), and then incubated at 37°C for 16–18 h. Beef meat samples (25 g) were homogenized in 10% NaCl-TSB (225 mL) and enriched at 37°C for 24 h, and then 10 μL samples were streaked onto Baired-Parker agar (BPA; Difco Laboratories) and cultured overnight at 37°C. Three presumptive S. aureus colonies per plate were selected and subcultured for identification. S. aureus isolates were identified using 16S rRNA sequencing and the Vitek 2 system (bioMérieux, Marcy-l'Étoile, France).
Susceptibilities to antimicrobial agents were determined according to the Clinical and Laboratory Standards Institute guidelines (CLSI, 2019). The antimicrobial agents tested were ampicillin (10), cefoxitin (30), chloramphenicol (30), ciprofloxacin (5), clindamycin (2), dalfopristin-quinupristin (15), erythromycin (15), gentamicin (30), mupirocin (200), rifampin (5), sulfamethoxazole-trimethoprim (23.73–1.25), and tetracycline (30). All antibiotic disks were obtained from BD BBLTM, except mupirocin, which was purchased from Oxoid (UK). Standard E-test (bioMérieux) assays were performed to determine minimum inhibitory concentrations (MICs) to vancomycin, oxacillin, tetracycline, and teicoplanin on Meuller-Hinton agar (MHA) plates.
The MICs to zinc chloride on all S. aureus isolates were determined by the standard agar dilution assay (ranging from 0.25 to 32 mM) in MHA II as reported before (Aarestrup and Hasman, 2004). Break point for zinc resistance was set at MIC of >2 mM according to the previous report (Aarestrup and Hasman, 2004). All susceptibility tests were repeated at least three times on separate days. The presence of the czrC (zinc resistance gene) was detected by PCR amplification as described before (Cavaco et al., 2010).
Multilocus sequence typing (MLST) was performed on the 5 MRSA and 11 MSSA isolates as described before (Enright et al., 2000). Briefly, seven target loci (arcC, gmk, tpi, aroE, glpF, pta, and yqiL) were PCR-amplified for subsequent sequencing analyses to determine the STs based on the MLST database (http://pubmlst.org/saureus/).
Types of SCCmec were determined on the 5 MRSA strains by multiplex PCR analysis as reported previously (Kondo et al., 2007). Multiplex PCR assays were performed to detect mec regulatory elements and cassette chromosome recombinase genes (ccrA1, ccrA2, ccrA3, ccrB1, ccrB2, ccrB3, ccrB4, and ccrC). The SCCmec types were determined based on the types of ccr and mec gene complexes.
The agr (I–IV) and spa types were determined for the MRSA and MSSA isolates, respectively, by PCR-based methods as described before (Gilot et al., 2002). For spa typing, the amplified genes were sequenced to determine the number of tandem repeats, and the spa type was assigned to each S. aureus strain according to the SpaServer database (http://spa.ridom.de/) (Harmsen et al., 2003). All MRSA and MSSA isolates were PCR-screened for the presence of tetK, tetL, tetM, tetO, and tetS genes, which confer resistance to tetracycline (Ng et al., 2001).
Multiplex-PCR analyses were performed on all MRSA and MSSA isolates to examine the presence of 19 SE genes and the tst1 gene (toxic shock syndrome toxin-1 gene) (Park et al., 2011). Briefly, four sets of multiplex PCRs were performed for the detection of the SE genes (PCR 1: sea, seb, sec, sed, and see; PCR 2: seg, seh, sei, sej, and sep; PCR 3: sek, sem, seo, and tst1; and PCR 4: sel, sen, seq, ser, and seu). Genomic DNA samples from the reference S. aureus strains were used for positive control PCR reactions (FRI913 strain for sea, sec, see, sek, seq, and tst1 ; FRI472 strain for sed, seg, sej, sel, sem, sen, seo, ser, and seu; MW2 strain for seh; COL strain for seb; and N135 strain for sei, and sep). Detection of Panton-Valentine leukocidin (PVL) genes, lukF and lukS, were performed by using a PCR-based method as reported previously (Lina et al., 1999).
Static biofilm formation assays were performed on all the MRSA and MSSA isolates as described previously (Pompilio et al., 2015). Briefly, staphylococci from overnight cultures were adjusted to OD600 of 0.1, diluted ~1:100 with fresh brain heart infusion (BHI) broth containing 30 mM glucose, and then 200 μL of the S. aureus culture was aliquoted to 96-well culture plates (SPL Life Sciences, Pochun, Korea). After 48 h incubation at 37°C, the culture supernatant was discarded, and then the plates were washed four times with phosphate-buffered saline. Next, the plates were air dried, and cells attached to the surface were stained with 5% safranin for 5 min. After the safranin staining, 30% acetic acid (Difco laboratories) was added to each well and absorbance was determined at OD492nm. A minimum of four independent experiments were performed on separate days.
Cationic antimicrobial peptides (CAPs) represent key elements in the host innate immune response. Porcine cathelicidin (PMAP-36) (Scheenstra et al., 2019) and human cathelicidin (LL-37) (Ouhara et al., 2008; Scheenstra et al., 2019) were synthesized with a purity of ≥95% (GL Biochen, Shanghai, China). In vitro survival assays against PMAP-36 and LL-37 were performed as described before using a 2 h microdilution method (Xiong et al., 2005). CAP susceptibility assays were performed with PMAP-36 (0.1 μg/mL) and LL-37 (3 μg/mL) using an initial S. aureus inoculum of ~5×103 CFUs in RPMI-1640 containing 5% LB broth. The concentrations of PMAP-36 and LL-37 were selected after extensive preliminary assays that showed sub-lethality with <50% reductions in CFUs over the 2 h time in both MSSA and MRSA strain groups. The data were analyzed and expressed as the % of relative mean survival±SDs of CAP-exposed versus CAP-unexposed controls. Three independent CAP susceptibility assays were performed for each MRSA and MSSA strain.
Results
Sixteen strains of MRSA (n=5, 0.39%) and MSSA (n=11, 0.86%) were cultured from 1,285 samples obtained from beef cattle farms, retail markets, and slaughterhouses over the twelve months of the sampling period (Table 1). The overall prevalence of MRSA was 0.5% in cattle farms and 0.3% in slaughterhouses. No MRSA isolates were recovered from beef meat, workers, or facility environment samples in retail markets. As shown in Table 1, four of the MRSA strains were recovered from cattle farms and one MRSA strain was cultured from a carcass sample in a slaughterhouse. Three of the five MRSA strains from beef cattle were isolated from the Gangwon province, and the two other MRSA strains were cultured from a farm worker and a carcass sample in Gyeongsang province (Table 1).
The overall prevalence of MSSA strain was 0.5% (n=4), 2.3% (n=4), and 0.9% (n=3) in cattle farms, retail markets, and slaughterhouses, respectively. Three of the MSSA strains were isolated from beef cattle, six MSSA strains were isolated from facility workers (one farm worker, one slaughterhouse worker, and four retail market workers), and two MSSA strains were isolated; one from a carcass and another from a slaughterhouse environment (Table 2). The geographic distribution of the 11 MSSA strains were as follows: Gyeonggi (n=4, 36.4%), Chungcheong (n=1, 9%), Jeolla (n=4, 36.4%), and Gyeongsang (n=2, 18.2%) provinces (Table 1).
MRSA, methicillin-resistant Staphylococcus aureus; MSSA, methicillin-susceptible Staphylococcus aureus; MLST, Multilocus sequence typing; MIC, minimum inhibitory concentrations; ST, sequence type; NT, non-typeable; AMP, ampicillin; CEF, cefoxitin; CIP, ciprofloxacin; CLI, clindamycin; ERY, erythromycin; TET, tetracycline; MUP, mupirocin; GEN, gentamicin; CHL, chloramphenicol; SYN, quinupristin-dalfopristin.
Seven different STs were assigned to the 16 S. aureus strains through MLST-analyses, with two non-typeable MSSA strains: BJFA-222 and BKFH-451 (Table 2). The three MRSA strains isolated from cattle were ST5 with SCCmec type II (ST5-SCCmec II), and two other MRSA strains each isolated from a farm-worker and a carcass sample were ST72-SCCmec type IV. The 3 ST5-SCCmec II MRSA strains were isolated from a single cattle farm and had identical spa type of t002 and agr type II. The two ST72-SCCmec IV MRSA strains had spa types of t664 and agr type of I.
As presented in Table 2, MSSA strains exhibited 6 different ST types: ST1 (n=3, 27.3%), ST188 (n=2, 18.2%), ST7 (n=1, 9.1%), ST72 (n=1, 9.1%), ST2199 (n=1, 9.1%), and ST2416 (n=1, 9.1%), and two non-typeable strains. Similar to the diverse MLST types, the 11 MSSA strains had 8 different spa types and two different agr types. Combined analyses of MLST, spa, and agr types in the MSSA strains revealed that the 11 MSSA strains were each genetically distinct, except the two ST1 MSSA strains, BSFA-4104 and BJSM-273. Although one ST72 MSSA strain (BJMH-426) and two ST72 MRSA strains (BKFH-321E and BKSM-133E) had identical MLST and agr types, differences in spa types were observed between the MSSA and MRSA strains (t664 vs. t126).
All the 16 S. aureus strains were susceptible to rifampin, sulfamethoxazole-trimethoprim, teicoplanin, and vancomycin. In contrast, the 5 MRSA strains and 7 of the 11 MSSA strains showed resistance to ampicillin (Table 2). Of note, the 5 MRSA strains displayed a multidrug resistance (MDR) phenotype by showing resistance to more than three antimicrobial agents tested. The ST72 MRSA-SCCmec IV strain, BKSM-133E, showed resistance to mupirocin, which is not used in animals. Similar to the MRSA strains, two of the MSSA strains, BSFA-4104 and BSSH-171, displayed an MDR phenotype. The BSSH-171 MSSA strain (ST2199) showed resistance to 7 different antimicrobial agents. In contrast to the two ST72 MRSA strains, the ST72 MSSA strain (BJMH-426) was susceptibility to all the antimicrobial agents tested.
PCR analyses of five tetracycline resistance genes (tetK, tetL, tetM, tetO, and tetS) revealed that the two ST72 MRSA strains and the ST2199 MSSA strain harbored tetM gene. Interestingly, although two of the three tetM-positive S. aureus strains (BKFH-321E and BSSH-171) exhibited a high level of tetracycline MICs (16 and 32 μg/mL, respectively), the BKSM-133E strain (ST72 MRSA strain) showed tetracycline MIC of 0.125 μg/mL (Table 1).
Resistance to zinc chloride has been proposed to be associated with specific genotypes of LA-MRSA isolates, such as CC398 MRSA with SCCmec V (Graveland et al., 2011). Although 3/5 MRSA (60%) and 8/11 MSSA (72.7%) strains were positive for czrC, all 16 S. aureus strains exhibited resistance to zinc chloride (MICs >2 mM) (Table 2). Of note, only czrC-positive strains displayed a high level of resistance (MICs ≥6 mM) to zinc chloride.
Although the five MRSA strains differed in MLST, SCCmec, agr, and spa types (ST5-SCCmec II-agr II-spa t002 vs. ST72-SCCmec IV-agr I-spa t664), all MRSA strains carried an identical set of SE genes (Table 3). The six SE genes (seg, sei, sem, sen, seo, and seu) detected in the five MRSA strains were within enterotoxin gene cluster (egc) 2 locus (Table 4). In contrast to the identical SE gene profile seen in MRSA strains, the eleven MSSA strains had ten different SE gene profiles. Interestingly, the ST72 MSSA strain (BJMH-426) had a set of SE genes that was identical to the two ST72 MRSA strains, suggesting a correlation between MLST type and SE gene profile.
2) Screened for the egc locus genes; seg, sei, sem, sen, and seo (Collery et al., 2009).
3) Screened for the egc locus genes; seg, sei, sem, sen, seo, and seu (Collery et al., 2009).
4) Screened for the egc locus genes; segvseiv, semv, senv, seov and seuv (Collery et al., 2009).
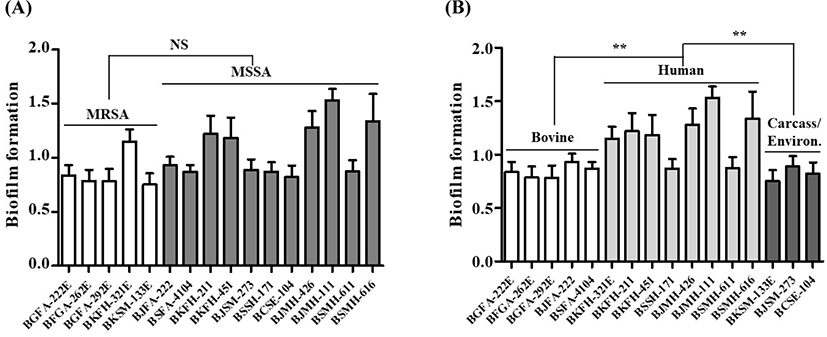
The formation of biofilm usually results in decreased susceptibility to antimicrobial drugs and host immune defense (i.e. antimicrobial peptide). Hence, the biofilm formation assays have been performed to assess virulence characteristics of the MRSA and MSSA strains in human and animal hosts (Abdelhady et al., 2014; Osman et al., 2016). As shown in Fig. 1A, ability to form biofilm under static condition was not different between MRSA and MSSA strains. However, as shown in Fig. 1B, in a comparison of S. aureus strains as groups of bovine-, human-, and non-bovine/non-human-associated strains regardless of methicillin-resistance, S. aureus strains from human hosts exhibited a significantly higher level of biofilm formation than the other two groups of S. aureus strains (p<0.01).
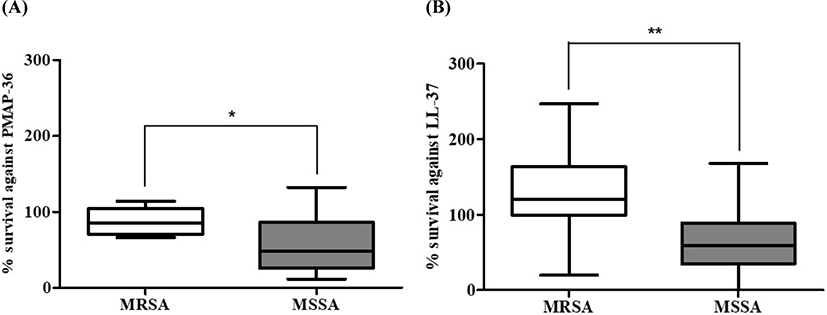
In vitro CAP susceptibility assays of all 16 strains revealed that MRSA strains tended to have significantly higher levels of resistance to PMAP-36 (p<0.05) and LL-37 (p<0.01) than MSSA strains. When the S. aureus isolates were compared as groups of bovine-, human-, and non-bovine/non-human-associated strains, no significant differences in susceptibilities were observed among the three strain groups (data not shown).
Discussion
Food-producing animals are well-known reservoirs for MRSA and MSSA, and an increasing number of cases have been reported in which humans are infected with S. aureus via foods of animal origin (Lim et al., 2010; Osman et al., 2016; Pauly et al., 2019). Recently, swine-associated MRSA has been reported worldwide; particularly, the ST398 MRSA in European countries (Graveland et al., 2011; Price et al., 2012; Vanderhaeghen et al., 2010). Although the presence and antimicrobial resistance of MRSA and MSSA have been investigated extensively in dairy cattle farms and beef meat samples (Lim et al., 2010; Osman et al., 2016; Song et al., 2016; Vanderhaeghen et al., 2010), few studies have been conducted on the occurrence and molecular composition of the MRSA and MSSA in beef cattle farms.
In this study, we investigated the occurrence and characteristics of MRSA and MSSA in the beef production chain, covering beef cattle farms, slaughterhouses, and meat samples in retail markets. Because MRSA and MSSA can be transmitted between livestock animals or beef meat and farmers or workers through frequent contact, samples were collected systematically from beef cattle, beef meat, humans, and facility environment. Overall, the prevalence of MRSA was 0.5% and 0.3% in cattle farms and slaughterhouses, respectively (Table 1). Several previous studies in Korea reported detection rates of 1.3%–13.9% for MRSA in bovine mastitic milk samples (Moon et al., 2007; Nam et al., 2011; Song et al., 2016). However, no previous studies have been conducted on the national prevalence of MRSA in beef cattle farms in Korea. The prevalence of bovine-originated MRSA in this investigation was lower than the prevalence rates reported in pigs from previous studies (2%–8.6%) in Korea (Lim et al., 2010; Moon et al., 2015). Weese et al. (2012) also reported the inability to isolate MRSA in a large group of beef cattle (491 nasal swabs and 488 fecal samples) in Canada. However, the prevalence of MRSA in beef cattle farms in Belgium was 10.16% (Nemeghaire et al., 2014), suggesting geographical variation. The overall prevalence of MSSA in cattle farm in this study was 0.5%, indicating that the occurrence of S. aureus in beef cattle farm is very low, regardless of methicillin resistance. Interestingly, as shown in Table 1, the isolation rates of MSSA in samples from slaughterhouses and retail markets (0.9% and 2.3%, respectively) were slightly higher than those of MRSA (0.3% and 0%, respectively). In a previous study, Kim et al. (2015) also reported isolation rates of MSSA (~6%) and MRSA (0.2%) in domestic beef meat samples. However, none of the MRSA and MSSA strains was isolated from beef meat samples in this study (Table 2).
The spread of MRSA in livestock and foods of animal origin has been linked to a high prevalence of CC398 LA-MRSA strains in the pig population and pork production system worldwide (Graveland et al., 2011; Moon et al., 2015; Price et al., 2012). Similarly, the most frequent swine-associated MRSA strain in Korea has been CC398 with t571 or t034 spa types (Moon et al., 2015). In contrast to the pork production chain, no CC398 genotype was observed among the MRSA and MSSA strains isolated from the beef production chain in this study (Table 2). Interestingly, the three MRSA strains from beef cattle were ST5 and two MRSA strains, one each from a farm-worker and a slaughterhouse carcass sample, were ST72, which are the most significant HA-MRSA and CA-MRSA strains in Korea (Kang et al., 2019). The three ST5 MRSA strains were isolated from different bovine hosts on a single farm and had identical genotypes (spa type of t002, SCCmec II, and agr type II) and antimicrobial resistance phenotypes, indicating colonization of beef cattle with a single clone of an ST5 MRSA strain. The two ST72 MRSA strains (BKFH-321E and BKSM-133E) each displayed resistance to three antimicrobial agents, while the ST5 MRSA strains exhibited an MDR phenotype to five antimicrobial agents (Table 2). A higher prevalence of MDR phenotype in ST5 MRSA vs. ST72 MRSA has previously been reported in Korea (Kang et al., 2019; Park et al., 2015). These data suggest that the two ST72 MRSA strains originated from human hosts rather than beef cattle. Unlike the MRSA strains, which displayed two major genotypes (ST5-SCCmec II-spa t002-agr II and ST72-SCCmec IV-spa t664-agr I), MSSA strains displayed diverse MLST, spa, or agr types (Table 2). Of note, the three MSSA strains from bovine hosts differed in their ST, spa, or agr types from the MSSA strains from human hosts, suggesting that transmission of MSSA from beef cattle to retail meat is still rare in Korea.
The high prevalence of CC398 LA-MRSA strains in swine farms seems to be associated with resistance to antimicrobial agents, particularly tetracycline compounds (Larsen et al., 2016; Moon et al., 2015; Price et al., 2012). In addition, zinc resistance conferred by the czrC has been proposed to be involved in the persistence of CC398 LA-MRSA strains in pigs (Aarestrup and Hasman, 2004; Cavaco et al., 2010; Price et al., 2012). As shown in Table 2, 3/5 MRSA strains (60%) and 8/11 MSSA strains (72.7%) harbored czrC, and a high level of zinc resistance was observed only in czrC-positive strains. However, due to the limited number of strains used in this study, a correlation between zinc resistance and other genotypic or phenotypic factors (e.g. MLST, SCCmec type, agr type, or origin of the strains) could not be determined. Although two ST72 MRSA strains and one ST2199 MSSA strain were positive for tetM, the BKSM-133E (ST72 MRSA) strain showed susceptibility to tetracycline (MIC 0.125 μg/mL) (Table 2).
Virulence factors may have played a crucial role in the prevalence and evolution of epidemic strains of MRSA and MSSA (Park et al., 2015). A variety of virulence-related factors have been identified in S. aureus strains, such as SE genes and toxic shock syndrome toxin-1 (TSST-1) (Hennekinne et al., 2012; Park et al., 2011; Park et al., 2015). These toxins are known to be pyrogenic and are often associated with human food poisoning and toxic shock syndrome (Park et al., 2011; Weese et al., 2012). As presented in Tables 3 and 4, all five MRSA strains possessed an identical set of seg, sei, sem, sen, seo, and seu genes that belongs to the egc2 locus (Collery et al., 2009). Similar to the ST5 and ST72 MRSA strains, the ST72 MSSA strain, BJMH-426, also carried the SE genes of egc2, suggesting a genetic correlation between ST72/ST5 and SE gene profiles.
The ability of S. aureus to adhere to extracellular adhesion molecules on host cells is an important virulence determinant for colonization in human and animal hosts (Osman et al., 2016). As shown in Fig. 1A, no significant difference was observed in biofilm formation between MRSA and MSSA strain groups (p=0.102). However, S. aureus strains isolated from human hosts tended to have a significantly enhanced level of biofilm formation versus strains from bovine hosts (Fig. 1B). These data, in combination with the genetic profiles of MRSA and MSSA strains, suggest that S. aureus strains need to orchestrate virulence factors such as biofilm formation to colonize new hosts and for successful transmission between human and animal hosts.
In addition to biofilm formation, S. aureus must overcome the bactericidal action of host antimicrobial peptides (Kang et al., 2019). Cathelicidins, small amphipathic peptides with a net positive charge, are abundant in host cells such as neutrophils and epithelial cells of the skin and upper respiratory tract (Kang et al., 2019). The persistence and successful colonization of the skin and upper respiratory tract would require S. aureus to resist the bactericidal action of cathelicidins. As presented in Fig. 2A and B, MRSA strains exhibited significantly higher levels of resistance to PMAP-36 (p<0.05) and LL-37 (p<0.01) than the MSSA strains. Although a difference in resistance to host antimicrobial peptides was observed in a small number of MRSA and MSSA strains, we speculate that the MRSA strains with MDR phenotype in combination with cathelicidin resistance can have an enhanced ability to survive on human and animal hosts under antimicrobial exposure.
We recognize that the present study has several limitations. Most data were resulted from a limited number of MRSA and MSSA isolates due to a very low prevalence of S. aureus in the samples collected in 2018. Information on the usage of antimicrobial agents and the concentration of zinc chloride in the animal feed in cattle farms were not available in this study. However, the current study is the first to systematically sample beef cattle, humans, and carcass/beef meat samples in different sectors of the beef production chain in Korea.
In conclusion, our data provide important information on the occurrence of MRSA and MSSA in the beef production chain in Korea. The prevalence of bovine-associated MRSA and MSSA was much lower than that of swine-associated MRSA and MSSA in Korea and other Asian and European countries. Unlike CC398 LA-MRSA in pigs, resistance to tetracycline and zinc chloride did not correlate with enhanced levels of MDR phenotype nor the prevalence of specific MLST types in bovine-associated MRSA and MSSA strains. In addition, increased levels of resistance to cathelicidins (PMAP-36 and LL-37) were observed in MRSA strains compared to MSSA strains. Although prevalence of MRSA is low in beef cattle in Korea, increased resistance to antimicrobial drugs and antimicrobial peptides may play an important role in the transmission of MRSA to humans via direct and/or indirect contact. Since livestock animals can serve as a reservoir for MRSA and MSSA, extensive surveillance for antimicrobial resistant pathogens and antimicrobial usage in the beef production chain is necessary to prevent potential transmission of the pathogens to humans.













