Introduction
Extracellular vesicle (EV) is a term for a vesicle released from cells to extracellular environment. EVs are found in a variety of prokaryotic and eukaryotic cells, and they are known to be responsible for the intercellular communications (Zaborowski et al., 2015). Both gram-positive and gram-negative bacteria secrete EVs, which carry proteins, nucleic acid, toxins, and cell wall components inside the 20–300 nm size of lipid bilayer cargo (Kim et al., 2015). Bacterial EVs recently have been spotlighted as key messengers of host-microbe interactions since it can deliver bacterial compounds directly into host cells in a stable and targeted form (Sanchez et al., 2010). Many recent studies have depicted that EVs are able to enter hosts’ epithelial cells and directly modulate host immunity (Sanchez et al., 2010). For example, it was reported that EVs secreted from Bifidobacterium bifidum LMG13195 helped differentiation of forkhead box protein 3 (Foxp3) 1 Treg cells, and Lactobacillus plantarum-derived EV components prolonged the survival of Caenorhabditis elegans which was exposed to vancomycin-resistant Enterococcus faecium (VRE) (Li et al., 2017; Lopez et al., 2012).
Family 5 extracellular solute-binding protein (ESBP) is one of the two components of the EV of Bifidobacterium longum KACC 91563. In general, ESBP is known to be capable of chemotaxis, transmembrane transport, and facilitating sensory transduction pathways (Tam and Saier, 1993). In our previous works, oral administration of the strain to a mouse model resulted in significantly decreased food allergy symptoms, and it also led to significant increase of Bifidobacterium and decrease of harmful bacteria such as Enterobacteriaceae and Clostridium and increased level of SCFA in feces when administered to healthy dog group (Kim et al., 2016; Park et al., 2018). It was also demonstrated that EVs produced by B. longum KACC 91563 were selectively internalized into mast cells and induced apoptosis of mast cells (Kim et al., 2016). The EV from B. longum KACC 91563 was identified to contain two proteins, ESBP and ASBP (ABC transporter, substrate-binding protein). Of these two proteins, only ESBP was capable of reducing the number of mast cells in the small intestine and ameliorating symptoms of food allergy (Kim et al., 2016). In particular, the ESBP had impacts specifically on the mast cells without suppressing any T cell-mediated immune responses.
Probiotic products were traditionally considered to exert proper health-promoting effects only when administered alive, but recent evidences are increasing that administration of the postbiotic molecules secreted from beneficial bacteria may be sufficient to promote the desired effects (Heo et al., 2018; Ruiz et al., 2014). Moreover, since administration of live bacteria strains always involves a risk of acting as a reservoir for antibiotic resistance genes which have potential to pass them to pathogenic bacteria and its stability and viability in host body is difficult to determine, utilization of probiotic bacterial extracellular molecules may represent a safer alternative way for the application of probiotic bacteria (Mehdi et al., 2016; Yan et al., 2007). For this reason, recently, there is a growing interest in the application of postbiotic molecules in the food products production. Postbiotic molecules from L. plantarum YML007 has been utilized as bio-preservative on soybeans grains, and nisin produced by Lactococcus lactis subsp. lactis strains is utilized in food products including canned soups, cheeses, mayonnaise, and baby foods, as a food preservative (Aguilar-Toala et al., 2018; Ahmad Rather et al., 2013; Chen et al., 2003). Postbiotic metabolites also have been shown to achieve high productivity and better health when used as animal feed additive (Loh et al., 2014). Utilization of postbiotic metabolites are not limited to functional foods. They have been introduced with potential pharmaceutical applications in the prevention or treatment of disease. For example, CytoFlora®, molecules from micronized cell wall lysates of several Lactobacillus and Bifidobacterium species, has been used to promote immune response and reduce intestinal dysbiosis in autistic children (Ray et al., 2010). Despite the vast application potentials like other well-known postbiotics, large-scale production of ESBP using recombinant DNA technology had not been explored before. Therefore, we report here the first study of cloning, expression, and purification of recombinant ESBP from B. longum KACC 91563 into E. coli strain BL21 (DE3).
Materials and Methods
All the enzymes for DNA cloning and amplification were purchased from TAKARA (Kusatsu, Japan), and the expression vector pET-28a(+) was from Invitrogen (CA, USA). E. coli strain BL21 (DE3) and BL21 (DE3) RIPL was also from Invitrogen (CA, USA). LB/Amp broth was from Difco (MD, USA), and isopropyl-beta-D-thiogalactopyranoside (IPTG) was purchased from Invitrogen (CA, USA). The Ni-NTA affinity matrix was purchased from QIAGEN (Hilden, Germany). Buffers and all other chemicals were purchased from Sigma-Aldrich (MO, USA).
B. longum subsp. longum KACC 91563 was provided by the National Institute of Animal Science, Rural Development Administration. It was originally isolated from feces of healthy Korean neonate in our previous study (Ham et al., 2011). E. coli BL21 (DE3) and E. coli BL21 (DE3) RIPL were used as recombinant protein expression host bacteria for gene cloning. pET-28a(+) was chosen to construct expression vector.
1,650 bp gene fragment encoding ESBP protein of 546 amino acid residues (Supplementary Information) was inserted into the histidine-tagged (His-tag) pET-28a(+) vector (Fig. 1), and then expressed in both E. coli strain BL21 (DE3) and BL21 (DE3) RIPL to attain optimized induction condition. Following enzyme digestion (NdeI/XhoI), the target DNA fragments were ligated into the NdeI/XhoI sites of pET-28a(+) vector to construct the fusion expression vector. Then the ligated product pET28a-KACC91563 was transformed into both E. coli strains.
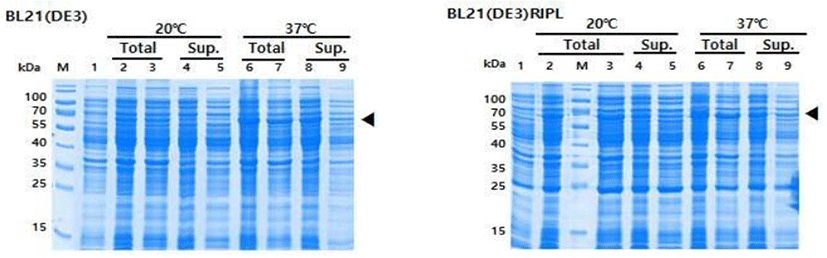
Both E. coli strains carrying the expression vector were grown in LB broth containing ampicillin (LB/Amp) at 20°C and 37°C for overnight with continuous shaking at 200 rpm to optimize the culturing and induction temperature. The culture was diluted 1:50 or 1:100 in a 5 mL of LB/Amp and incubated until the OD600 reached 0.4–0.5. Protein expression was induced by adding IPTG to a final concentration of 1.0 mM and incubation was continued at 37°C with shaking at 200 rpm, and 0.5 mL of fractions were collected every hour for 3 h. Also, 1 mL was taken from the culture to make a second tube simultaneously after adding IPTG, and it was incubated at 20°C with shaking at 200 rpm as well for 16 h, and then 0.5 mL of fraction was used for analysis. The cells were harvested by centrifugation and resuspended in 50 uL of SDS sample buffer. The sample was boiled for 3 min and examined for the position of expressed protein by SDS-PAGE. 12% SDS-PAGE gel was loaded with 5 μL (equivalent to 100 μL of cells) and the bands were visualized by Coomassie brilliant blue.
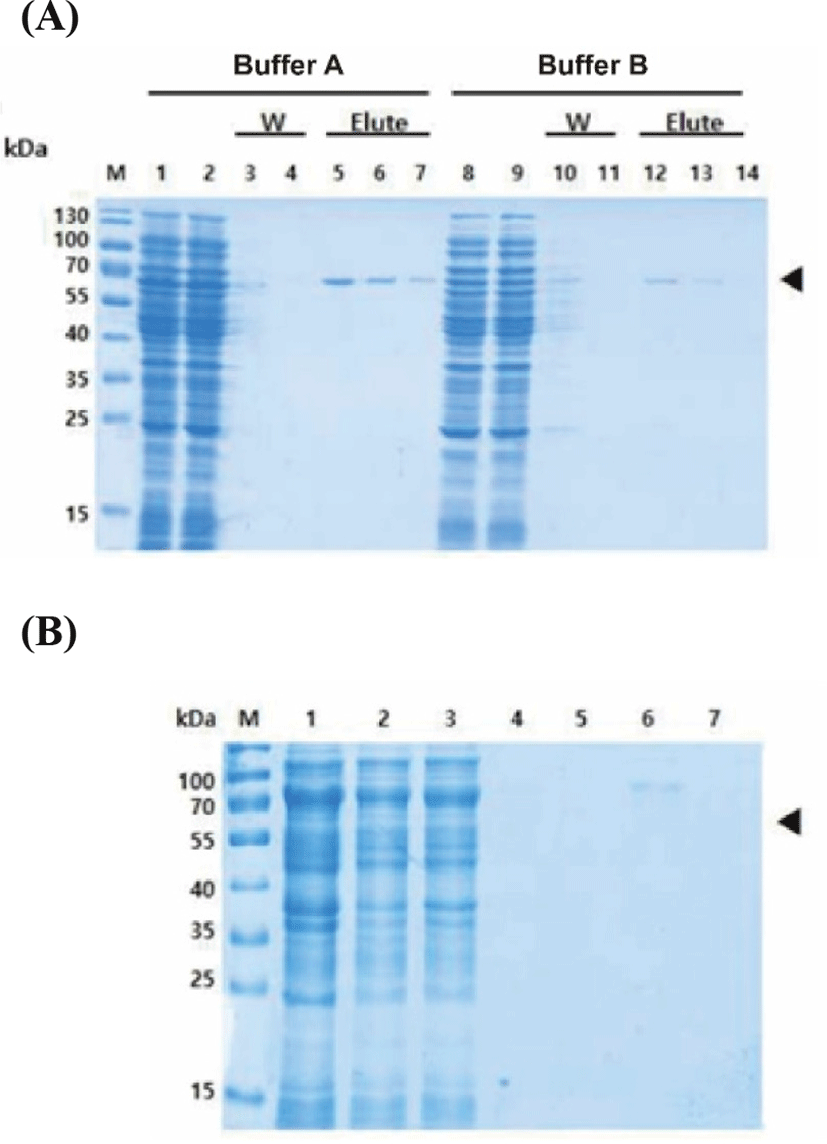
ESBP protein was purified via affinity chromatography using a nickel-nitrilotriacetic acid (Ni-NTA) gel matrix. For optimization purpose, purification was performed under both native condition and denatured condition with different sets of buffers (Table 1). The harvested cells were resuspended in each type of Ni-NTA lysis buffer (Table 1) by vigorous poking, stirring, and pipetting. The resuspended cells were lysed by sonication on setting 3, in an ice bucket, pulsed with 30 sec on, 30 sec off for 5 min total. To clarify the cell lysate, additional sonication on setting 4 were done for 5 min. The lysates were transferred to a column containing Ni-NTA resin. The column was washed with buffer (Table 1). The protein was then eluted by increasing the imidazole concentration to 250 mM. SDS-PAGE and Coomassie blue staining was used to analyze the purity of the ESBP following the purification process.
Results
To examine the growth profile and the expression level of recombinant ESBP protein, pET281-KACC91563 was transformed into two E. coli strains BL21 (DE3) and BL21 (DE3) RIPL and used for protein induction and expression. Each transformed strain was also grown and induced separately at 20°C, 37°C to explore the optimal temperature. We performed SDS-PAGE analysis to check protein size and optimize expression. As shown in Fig. 2, expression of ESBP protein from lysate supernatant was successful in both E. coli strains, but the expression was improved when the culture was incubated and induced at 37°C. Therefore, it can be concluded that 37°C is the optimal temperature for producing ESBP protein.
To determine the suitable purification protocol for ESBP protein, purification was carried out separately under native and denaturing conditions using the Ni-NTA column. One sample of an induced culture was lysed under native condition while another sample was proceeded in a buffer containing 8 M urea. Coomassie-stained SDS-PAGE analysis of each sample collected through the purification steps is shown in Fig. 3. The presence of specific 60.0 kDa target protein band was confirmed with the native lysate (Fig. 3A), but with purification under denaturing condition, no visible protein band of the appropriate molecular weights eluted (Fig. 3B). The efficiency of the different buffer sets on purification under native condition was also tested (Table 1). As shown in Fig. 3A, the recombinant proteins were eluted with both buffer A and B, but the use of buffer A enhanced protein purification. Thus, it was determined that applying native condition using buffer A would be appropriate for the large-scale purification.
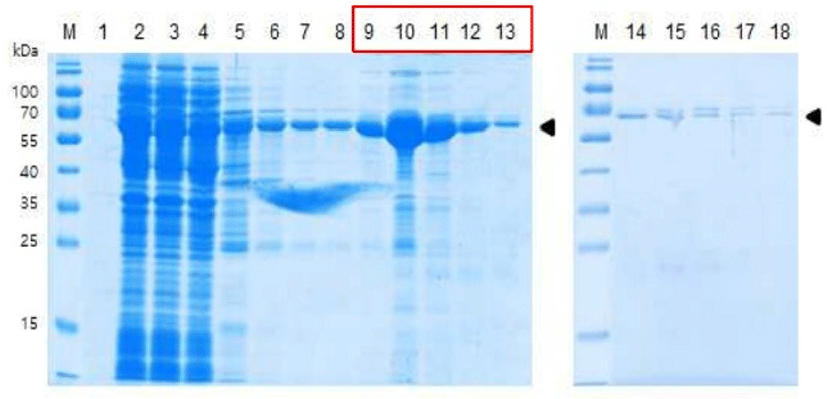
Based on the results from the small-scale culture optimization, the pET281-KACC91563 was transformed into E. coli strain BL21 (DE3), and the cells were grown at 37°C in LB broth containing ampicillin to a cell density of OD600=0.55–0.6. Expression was induced by adding 1 mM IPTG at 37°C as well, and then protein purification was performed under native condition with buffer A. For the purification, the cell pellet was dissolved in binding buffer (20 mM Tris, 10 mM NaCl, 5 mM imidazole (pH 8.0)), and the lysate-Ni-NTA mixture was loaded into Ni-NTA column. After the column had been washed with buffer (20 mM Tris, 10 mM NaCl, 20 mM imidazole (pH 8.0)), the His-tagged recombinant protein was eluted with elution buffers (20 mM Tris, 10 mM NaCl, 100 mM imidazole (pH 8.0) / 20 mM Tris, 10 mM NaCl, 250 mM imidazole (pH 8.0)). As a result, the elution of 60.0 kDa target ESBP protein band was confirmed (Fig. 4). Total 7.15 mg of purified protein was obtained from 2 L of culture with yield of 3.58 mg/L.
Discussion
The therapeutic potential of EVs from eukaryotic cells (e.g. mesenchymal stem cells) has gained increasing interests in recent years, and several industrial cell manufacturers have already been giving efforts to eukaryotic EVs production for clinical use (El Andaloussi et al., 2013; Rani et al., 2015). Yet, the research into the applications of bacterial EVs is still in its beginning stage to be investigated. However, bacteria also secrete many different types of EV containing proteins which perform diverse biological processes in the environment. Especially, a considerable amount of evidences has suggested in recent studies that the extracellular molecules produced by probiotic bacteria are linked directly to the health-promoting effects attributed to the corresponding strain (Sanchez et al., 2010). Several postbiotic molecules and its host immunomodulatory activity have been identified, and these findings provide opportunities for the development of new promising functional foods. EVs can be directly produced from large-scale cultures of bacteria in a straight forward way (Bitto and Kaparakis-Liaskos, 2017). However, the heterogenous nature of EVs makes it difficult to ensure consistency which is important for commercial application, and some EVs may even contain undesirable components like toxins (Bitto and Kaparakis-Liaskos, 2017). In addition, the presence of some EV-associated immunogens can rather induce adverse immune responses. To overcome these challenges, the development of large-scale production strategy targeting for specific EV protein of interest using prokaryotic expression systems is suggested. We report here the successful cloning and purification of the B. longum EV-derived protein, ESBP in E. coli, which was revealed to alleviate food allergy symptoms in our previous study. This is the first study of ESBP of probiotic bacteria production using recombinant DNA technology.
Currently most treatments for allergic diseases concentrate on either preventing the effects of histamine or inhibiting the body’s overall immune responses using various substances. Though, none of these attempts are completely effective, and suppression of overall immune responses can lead to another significant health problem. Since the ESBP has the specificity for mast cells, it will be very useful for the development of safe and effective alternative therapeutics. In this study, we produced the ESBP protein by cloning a gene encoding ESBP from B. longum KACC 91563 into E. coli BL21 (DE3). To enhance the efficiency of the protein purification, we have investigated and optimized three parameters which are the host bacterial strain, the culturing and induction temperature, and the purification protocol. It was confirmed that the expression level was improved when the culture was incubated and induced at 37°C. We could produce the recombinant protein under native condition, while no visible protein band of the appropriate molecular weights was observed with denaturing condition. Purification under native conditions is an ideal condition to preserve its biological activity and avoid significant loss of produced protein during the renaturation step (Saffarian et al., 2016). After the parameters affecting the amount of purified protein were explored and optimized, total of 7.15 mg of ESBP were produced from two liters of transformed culture. The production yield presented here might be not sufficient for mass production yet, but application of more recently devised protocols may facilitate increasing efficiency and yield production. Considering the particular beneficial effects of it, commercialization of the recombinant ESBP will have a potential to treat a wide range of allergic disease, which may have advantages over current therapeutic approaches and also can be used as a functional ingredient for production of various functional foods. We expect that the results presented in this study will serve as a starting point for further studies to make it possible.













