Introduction
Research area of human gut microbiota has been booming since gut bacteria are closely related to human physiology including immunity, obesity, diabetes, metabolic syndrome, brain health, and so on (Allaire et al., 2018; Rajani and Jia, 2018; Sun et al., 2018; Tengeler et al., 2018). In particular, human gut bacterial composition is one of the most important factors for maintaining intestinal homeostasis (Zhang et al., 2018). Balance between beneficial bacteria and harmful ones should be considered when measuring the health status of individual gut. Bifidobacteria and lactic acid bacteria (LAB) are among the most beneficial bacteria in our gut (Linares et al., 2017). Bifidobacteria are normally living in the colon. They produce not only lactic acid, but also acetic acid which shows strong antimicrobial activity against pathogenic bacteria (Fukuda et al., 2011; Tejero-Sariñena et al., 2012). These bacteria also possess other beneficial effects such as immunological, anti-diarrheal, anti-constipation, anti-inflammation, and anti-cancer effects (Alard et al., 2018; Escribano et al., 2018; Kang et al., 2017; Ruiz et al., 2017). For these reasons, bifidobacteria are broadly used as probiotics in functional food market (Kleerebezem and Vaughan, 2009). In particular Bifidobacterium catenulatum which belongs to the B. adolescentis group has been rarely studied but abundant in feces from humans (Felis and Dellaglio, 2007; Matsuki et al., 2004). Therefore B. catenulatum was used as a model for genetic recombination.
Molecular imaging is a powerful tool to investigate phenomena in biological nature. It gives researchers direct evidences via visualization (Ogawa and Takakura, 2018). In general, green fluorescence protein (GFP) and luciferase genes are used for labelling LAB and bifidobacteria. These labelled bacteria have been applied in in vitro, in situ, and in vivo tests to monitor their behaviors (Drouault et al., 1999; Grimm et al., 2014; Karimi et al., 2016; Lee et al., 2007; Moon and Narbad, 2017). The luxA-luxB gene cassette of Vibrio harveyi and the gfp gene of Aequora victoria were used for labeling Lactococcus lactis strain to investigate its survival, physiology, and lysis in the different compartments of murine digestive tracts (Drouault et al., 1999). The strain orally administered with the diet was resistant to gastric juice (90 to 98% survival), whereas 10-30% was survived in the duodenum. A Lactobacillus plantarum strain was labeled with a luciferase gene and monitored in a complex food matrix during fermentation (Moon and Narbad, 2017). The strain was successfully detected in the matrix during the fermentation by measuring bioluminescent signals. In a previous paper for Bifidobacterium sp., researchers have shown that a niche environment of a B. breve strain in murine intestine is the cecum via molecular imaging technique using a luciferase (Cronin et al., 2008).
In this study, we developed a novel labelling system for bifidobacteria using a luciferase gene based on a replicon of plasmid pFI2576 previously identified from B. longum FI10564 (Moon et al., 2009). The recombinant B. catenulatum [pTG262::pFI2576 rep (luc+)] successfully showed a luminescence signal.
Materials and Methods
Escherichia coli DH5α and E. coli MC1022 were cultured in LB broth (10 g/L NaCl, 10 g/L tryptone, and 5 g/L yeast extract, pH 7.0) at 37°C with shaking. B. catenulatum #27, which was previously isolated from human fecal samples in our lab and identified by 16S rRNA gene sequence analysis, was cultured in MRS (De Man, Rogosa, and Sharpe) broth at 37°C under anaerobic condition (Whitley A95 Anaerobic Workstation, Don Whitley Scientific, Bradford, UK). For antibiotic selection of recombinant strains, 100 μg/mL of ampicillin (Sigma, Poole, UK) was used for E. coli while 15 and 5 μg/mL of chloramphenicol (Sigma) were used for E. coli and B. catenulatum, respectively.
Plasmids and primers used in this study are listed in Table 1. pFI2576 rep (~2.0 kb) PCR product was produced with pFI2576 rep F-1 and R-1 primer set under the following PCR conditions: initial denaturation at 98°C for 30 s; 30 cycles of 98°C for 10 s and 72°C for 1 min; final extension at 72°C for 5 min. Phusion-HF DNA polymerase (Thermo Scientific, UK) was used for PCR. PCR product was digested with SalI restriction endonuclease (New England Biolabs, Hitchin, UK) and inserted into pUC19 cloning vector digested with the same restriction enzyme. A luciferase gene (luc+) was liberated with XbaI and EcoRI restriction endonucleases (New England Biolabs) from pLuc2 recombinant plasmid and inserted into pTG262 vector. The cloned pFI2576 rep was digested with SalI enzyme and subcloned into recombinant pTG262 (luc+) digested with the same enzyme. The final recombinant DNA was named pTG262::pFI2576 rep (luc+).
| Plasmid or primer | Description or sequence (5’→ 3’) | Reference |
|---|---|---|
| Plasmid | ||
| pFI2576 | 2.2 kb; A cryptic plasmid isolated from Bifidobacterium longum FI10564 | Moon et al., 2009 |
| pUC19 | 2.7 kb; E. coli cloning vector, Apr | Sambrook and Russell, 2001 |
| pUC19::pFI2576 rep | 4.7 kb; pFI2576 replicon (~2.0 kb) PCR product was inserted into pUC19, Apr | This study |
| pTG262 | 5.6 kb; Shuttle vector, Cmr | Fernandez et al., 2009 |
| pLuc2 | 8.5 kb; Shuttle vector harboring a luciferase gene (luc+), Apr, Emr | Eom et al., 2015 |
| pTG262 (luc+) | 7.4 kb; luc+ gene was inserted into pTG262, Cmr | This study |
| pTG262::pFI2576 rep (luc+) | 9.4 kb; pFI2576 rep was inserted into pTG262 (luc+), Cmr | This study |
| Primer | ||
| pFI2576 rep F-1 | acgcgtcgacggttgttgctccagcacc | This study |
| pFI2576 rep R-1 | acgcgtcgacctccaatcgtccgtcga | This study |
Transformation method was based on a previously published paper (Argnani et al., 1996). For preparation of electro-competent cells, 50 mL of B. catenulatum #27 were cultured in MRS broth supplemented with 0.05% L-cysteine and 0.5 M sucrose. After reaching OD600 of 0.31, they were set on ice for 20 min. These cells were washed twice with 45 mL of ice-cold electroporation buffer (0.5 M sucrose in 1 mM HEPES buffer, pH 6.0) and then washed with 1 mL of the same buffer twice. Cells were finally resuspended with 200 μL of the same buffer. Recombinant DNA of pTG262::pFI2576 rep (luc+) (105 ng) was transferred to 80 μL of electro-competent cells of B. catenulatum #27 under electroporation condition of 2.0 kV, 25 μF, and 200 Ω using GenePulser Xcell (Bio-Rad), recovered with 800 μL of MRS broth supplemented with 0.05% L-Cys and 0.5 M sucrose in an anaerobic cabinet (Whitley A95 Anaerobic Workstation), and selected on MRS agar plate supplemented with 5 μg/mL of chloramphenicol (Sigma).
Bioluminescence of recombinant E. coli and B. catenulatum was measured with a luciferase assay system (E1500; Promega, USA) according to manufacturer’s guidance. Cell culture (200 µL) was centrifuged, resuspended with 20 μL of phosphate buffered saline (Sigma), and loaded into a 96-well microplate (Black Costar flat bottom, Corning, USA). Luciferase assay reagent (100 µL) was added to the microplate and bioluminescent signal was measured with a BMG FLUOstar OPTIMA Microplate Reader (BMG LABTECH GmbH, Germany).
Results and Discussion
For construction of E. coli-Bifidobacterium shuttle vector, a Bifidobacterium plasmid replicon from pFI2576 was amplified by PCR. pFI2576 rep F-1 and R-1 primer set (Table 1) successfully amplified a PCR product with expected size of 2,012 bp (Fig. 1A). Partial nucleotide sequence of the PCR product was 100% identical to original pFI2576 replicon sequence (data not shown), indicating that PCR product was correctly amplified.
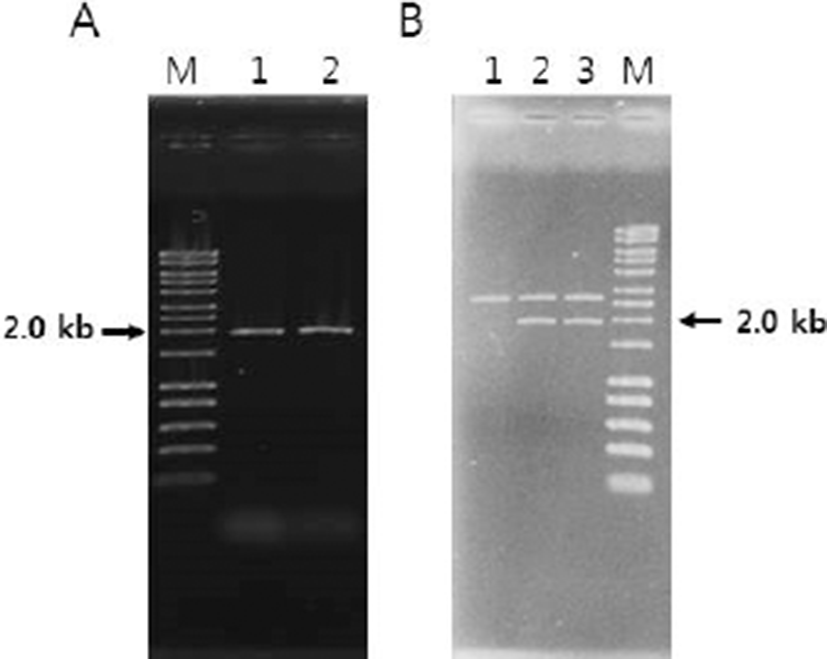
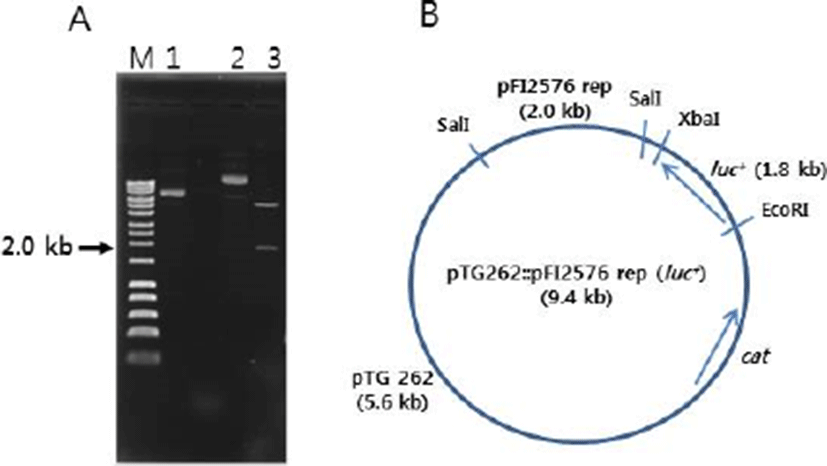
The pFI2576 replicon PCR product was inserted into pUC19 cloning vector. The insert (~2.0 kb) was confirmed by SalI restriction endonuclease digestion (Fig. 1B). The recombinant DNA pTG262 (luc+) was also confirmed by restriction digestion with EcoRI and XbaI endonuclease where luc+ gene (~1.8 kb) was liberated (Fig. 2A) from pTG262. pTG262::pFI2576 rep (luc+) was finally constructed. Its physical genetic map is shown in Fig. 2B.
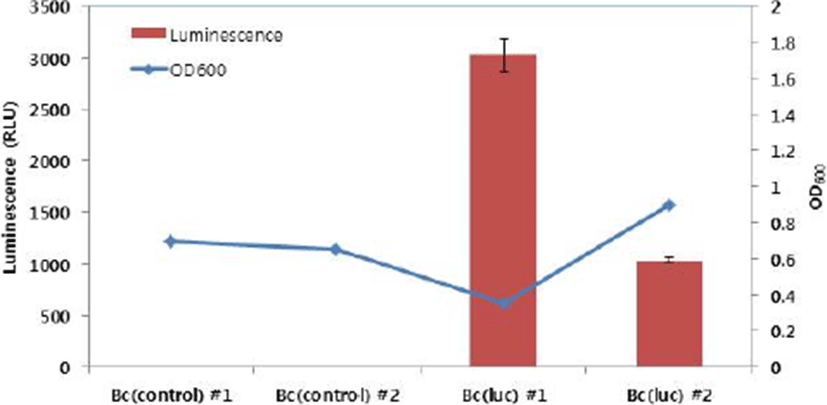
As mentioned above, pTG262::pFI2576 rep (luc+) was successfully constructed for labelling Bifidobacterium species. Before transforming Bifidobacterium sp., luminescent signal of E. coli MC1022 [pTG262::pFI2576 rep (luc+)] was measured. E. coli MC1022 [pTG262::pFI2576 rep (luc+)] showed very strong luminescent signal (1.0×106 RLU), whereas E. coli MC1022 (pTG262::pUC19::pFI2576 rep) as a negative control showed only basal signal (13 RLU), indicating that the recombinant DNA was properly constructed for bioluminescent labelling. These two recombinant DNAs were transferred to B. catenulatum #27. Transformation efficiencies for pTG262::pUC19::pFI2576 rep and pTG262::pFI2576 rep (luc+) were 1.3×103 and 4.8×101 CFU/μg DNA, respectively. Colonies (two each) were selected and cultured under anaerobic condition and luminescent signals were measured. As shown in Fig. 3, two cultures for B. catenulatum #27 (pTG262::pUC19::pFI2576 rep) presented luminescent signals of 2 and 4 RLU, whereas B. catenulatum #27 [pTG262::pFI2576 rep (luc+)] presented signals of 3,024 and 1,031 RLU. The difference in luminescent signal between the two cultures of B. catenulatum #27 [pTG262::pFI2576 rep (luc+)] might be due to growth state.
To date, very limited information is available for development of luciferase-based labelling system for bifidobacteria. Cronin et al. (2016) have introduced in vivo bioluminescence imaging technique where E. coli and B. breve are engineered to express lux gene cassette. Ninomiya et al. (2013) have used an imaging analyzing method to evaluate in situ bioluminescence from a recombinant B. longum expressing bacterial luciferase to understand the effect of dissolved oxygen concentration. Guglielmetti et al. (2008) have applied a bioluminescent labelling tool to study the physiological state of B. longum strain under various environmental conditions. As described in these papers, bioluminescent labelling system could be broadly used to understand physiological states and behaviors of strictly anaerobic Bifidobacterium sp. in in vitro, in situ, and in vivo. In the present study, we developed a luciferase-based labelling system for bifidobacteria. This system successfully worked in a B. catenulatum strain as a model. In the near future, the system could be applied to other Bifidobacterium species to investigate their physiology and roles under various environments.













