Introduction
In the food industry, Listeria monocytogenes is a serious concern because it is a human pathogen and causes the rare but life-threatening disease listeriosis. Most cases of Listeria infection are caused by cheeses and ready-to-eat meats (Cartwright et al., 2013; Garrido et al., 2010). Post-processing cross-contamination occurs owing to the presence of pathogens on equipment and in the environment in processing plants (Ferreira et al., 2014; Meloni et al., 2014; Ortiz et al., 2014). L. monocytogenes can grow at low temperatures (4°C to 10°C) that generally repress pathogen growth, and has a tolerance to acidic conditions (low pH) and high salt concentrations (Ferreira et al., 2014). In unborn fetuses, newborn babies, and elderly, pregnant, and immunocompromised adults, listeriosis occurs via L. monocytogenes invasion through the intestinal tract and a resultant systemic infection and is associated with a high mortality rate of up to 30% (Vázquez-Boland et al., 2001). In recent years, the number of cases of foodborne disease increased in Europe, and the increase was correlated with the incidence of listeriosis, especially in elderly people (≥ 65 years of age) (Goulet et al., 2008).
Antibiotics are used to treat bacterial illnesses; however, alternatives to antibiotics are in high demand because of the development of antibiotic resistance. Bacteriophages can be used as an alternative to antibiotics in the control of pathogens (Jassim and Limoges, 2014). Bacteriophages are extensively distributed in all environments and can kill bacteria, so they naturally play a role in controlling bacterial populations (Hagens and Loessner, 2014; Rodríguez-Rubio et al., 2015). Bacteriophages are thought to be safe for humans, animals, and plants (Rodríguez-Rubio et al., 2015). Biocontrol strategies based on bacteriophages have received an increasing amount of interest owing to their efficacy, practicability, safety, and economic feasibility (Farber and Peterkin, 1991; Hagens and Loessner, 2007; Hagens and Loessner, 2010; Hagens and Offerhaus, 2008). In fact, bacteriophage products for phage therapy and phage-based pathogen detection for the food and agriculture industries are being made commercially available (Schmelcher and Loessner, 2014; Sulakvelidze, 2013). For example, ListexTM P100 and ListShiedTM have been developed to combat Listeria monocytogenes and are commercially available after being deemed Generally Regarded As Safe (GRAS) by the FDA and USDA (Hagens and Loessner, 2014).
The objective of this study was to isolate bacteriophages that target L. monocytogenes, to characterize the isolated bacteriophages, and to evaluate their lytic activity against L. monocytogenes. In this study, listeria bacteriophages were isolated from fecal samples of broiler chicken raised at a poultry farm at Chung-Ang University, South Korea. Host specificity was evaluated with four L. monocytogenes strains. We also examined the morphological characteristics of the phages by transmission electron microscopy. The lytic activity of selected bacteriophages was further evaluated in liquid milk to investigate their potential use in dairy products.
Materials and Methods
Isolation of bacteriophages that target Listeria monocytogenes
Fresh six fecal samples from Hyline Brown laying hens (6 wk old) were collected from a poultry farm at Chung-Ang University, South Korea. Samples were immediately maintained on ice and stored at −80°C in a deep freezer. Pre-enriched samples were amplified using the doublelayer method for propagation of phages (Zinno et al., 2014). The double-layer method was conducted with tryptic soy agar (TSA) containing 1.5% agarose and semi-solid TSA containing 1.25 mM of CaCl2 and 0.5% agarose. Individual double-layered plates were inoculated with 0.1 mL of each of four L. monocytogenes strains (ATCC 7644, ATCC 15313, ATCC 19114, and ATCC 19115). After incubation at 25°C for 24 h, plaque was extracted from the plates. The recovered plaque was put into 1 mL of saline magnesium (SM) buffer (pH 7.5) in a 1.5 mL Eppendorf (EP) tube and was left overnight at 4°C. Then, the EP tubes were centrifuged at 10,000 g for 10 min at 4°C and the supernatant was propagated by the double-layer method as described previously. Propagated bacteriophages were collected with 4 mL of SM buffer (Carvalho et al., 2010; Janež and Loc-Carrillo, 2013; Salama et al., 1989). Bacteriophages titer was determined by the soft agar overlay method. The isolated bacteriophages were serially diluted in SM buffer and 0.01 mL of each was dropped on the surface of double-layered media covered with the L. monocytogenes host strains.
Host specificity of the isolated bacteriophages was determined by the double-layer method using four different serotypes of L. monocytogenes (Table 1). The plates were incubated at 37°C for 24 h. Host range was determined by the presence of a clear lysis zone on the plate (Zinno et al., 2014).
Four select bacteriophages (LMP1, LMP7, LMP8, and LMP12) were examined with L. monocytogenes ATCC 7644 or ATCC 19114. TSB (0.9 mL) was inoculated with 0.05 mL of an overnight culture of L. monocytogenes ATCC 7644 or ATCC 19114 to reach 5×105 CFU/mL in a 1.5 mL EP tube. Diluted bacteriophage (0.05 mL) at multiplicity of infection (MOI) values of 10 and 100 was added to the tubes. A culture containing 0.05 mL of SM buffer instead of bacteriophage was used as a negative control. The suspensions were incubated at 30°C for 24 h. Samples (0.2 mL of suspensions) were inoculated in 96-well plates in triplicate and optical density (O.D.) values at wave length of 600 nm were measured using a spectrophotometer (Epoch, BioTek, USA) at 2-h intervals after 10 h of incubation. The lytic ability of two bacteriophages (LMP1 and LMP7) in liquid media was also evaluated at 10°C with L. monocytogenes ATCC 7644 or ATCC 19114 (Albino et al., 2014).
One hundred milliliters of suspensions containing each of LMP1 and LMP7 were concentrated by ultracentrifugation at 320,000 g for 3 h using a Beckman L90K centrifuge equipped with a type 70 Ti rotor. The centrifuged pellet was suspended in 1 mL SM buffer to obtain 1010-1011 PFU/mL of phages. Bacteriophages were deposited onto carbon-coated copper grids and stained with uranyl acetate (2%, pH 4.5). Morphological observation was conducted at 80 kV with a transmission electron microscope (TEM). Morphological analysis of the bacteriophages was done with the Image J program (Denes et al., 2014).
The lytic ability of LMP1 and LMP7 was examined with L. monocytogenes ATCC 7644 and ATCC 19114 in milk. Milk (0.95 mL) was inoculated with overnight-cultured L. monocytogenes ATCC 7644 and ATCC 19114 (0.025 mL) to reach 5×105 CFU/mL in a 1.5 mL EP tube. Diluted bacteriophage (0.025 mL) at MOI values of 10 and 100 was added to the tubes. A culture with 0.025 mL of SM buffer was used as a negative control. Suspensions were incubated at 30°C for 20 h. The number of L. monocytogenes was counted using the plate culture method on Oxford agar (Becton, Dickinson and Company, USA) at 4-h intervals after 8 h of incubation (Albino et al., 2014).
Phages LMP1 and LMP7 were also evaluated at 10°C with L. monocytogenes ATCC 7644 or ATCC 19114 in skim milk. The evaluation test used was the same as that for the suspensions incubated at 30°C. The specimens were incubated at 10°C for 5 d. The number of host bacteria was counted daily using the plate culture method on Oxford agar (Albino et al., 2014).
Each experiment was measured in triplicate. Data from each group were expressed as mean and standard deviation. Statistical significance was analyzed at p<0.05 by one-way analysis of variance (ANOVA) and paired t-test using Statistical Package for Social Science (SPSS) software v23 (IBM, USA).
Results and Discussion
From the first stage of isolation, twelve bacteriophages (LMP1 to LMP12) against Listeria monocytogenes strains were isolated from chicken feces. Phage LMP1 was isolated with L. monocytogenes ATCC 7644, and the other eleven phages were isolated with L. monocytogenes ATCC 19115 as their hosts. The plaques of each phage isolate were selected and further purified by the soft agar overlay method, and each purified phage was successfully propagated at a titer up to 109-1011 PFU/mL with their original host strains.
The host specificity of the 12 bacteriophages were determined using four strains of L. monocytogenes (Table 1). Regardless of the serotypes of their host bacteria, all bacteriophage isolates showed lytic activity against L. monocytogenes ATCC 7644, ATCC 15313, ATCC 19114, and ATCC 19115. However, none of the phages showed lytic activity against L. monocytogenes ATCC 19111 (data not shown); further study will be needed to understand how this strain is resistant to the bacteriophages and what underlying resistance mechanisms the strain possesses. Host-specific lytic activity of a bacteriophage against its host bacteria is partly based on the interactions between the tail fiber of phages and the receptor components on the bacterial cell wall (Eugster et al., 2011). Also, bacteriophage endolysin is a hydrolyzing enzyme that targets the peptidoglycan moiety in the cell wall of host bacteria. The cell wall-binding domain in the C-terminal end of endolysin is responsible for the recognition of specific cell wall components. Also, the bacterial cell wall structure was determined by the presence of several genes. Even though genetic variation in L. monocytogenes serotypes remains to be further investigated (Eugster et al., 2011), different serotypes of this bacteria have different wall teichoic acid structures and glycosylation patterns. However, high lytic activity of the isolated bacteriophages against the four different serotypes of L. monocytogenes (Table 1) was confirmed by soft agar overlay, suggesting a potential use of these phages against a wide range of L. monocytogenes strains.
From the first twelve isolated bacteriophages, four phages (LMP1, LMP7, LMP8, and LMP12) and two host strains (L. monocytogenes ATCC 7644 and L. monocytogenes ATCC 19114) were selected for further study based on lytic activity patterns and host specificity. The lytic activity of the phages against the host strains was evaluated by measuring the optical density (OD) of the liquid medium during the growth of host bacteria at 30°C with different MOI (multiplicity of infection; the ratio of phages to their host cell) values (Fig. 1). Overall, the growth of L. monocytogenes strains was inhibited in an MOI-dependent manner in the same phage-host combinations (Figs. 1A vs. 1C, and 1B vs. 1D). The growth of L. monocytogenes ATCC 7644 was efficiently inhibited for up to 12 h (p<0.001) by LMP1 and up to 16 h (p<0.001) by LMP 7, LMP 8, and LMP 12, and growth recovered slowly thereafter, reaching the same OD values as seen in the untreated control after 24 h of incubation (Figs. 1A and 1C).
Completely different inhibition patterns were observed when L. monocytogenes ATCC 19114 was used as a host with the same phages. At an MOI of 10, the growth of L. monocytogenes ATCC 19114 was slightly delayed by LMP7, LMP8, and LMP12, whereas no growth was observed in an incubation of up to 14 h by LMP 1 (p<0.001) (Fig. 1B). Inhibition of the growth of L. monocytogenes ATCC 19114 was more prominent at an MOI of 100. LMP8 and LMP 12 were able to inhibit the growth of L. monocytogenes ATCC 19114 for up to 18 h (p<0.001) and LMP 1 and LMP 7 exhibited complete inhibition activity even after a 24-h incubation (p<0.001) (Fig. 1D).
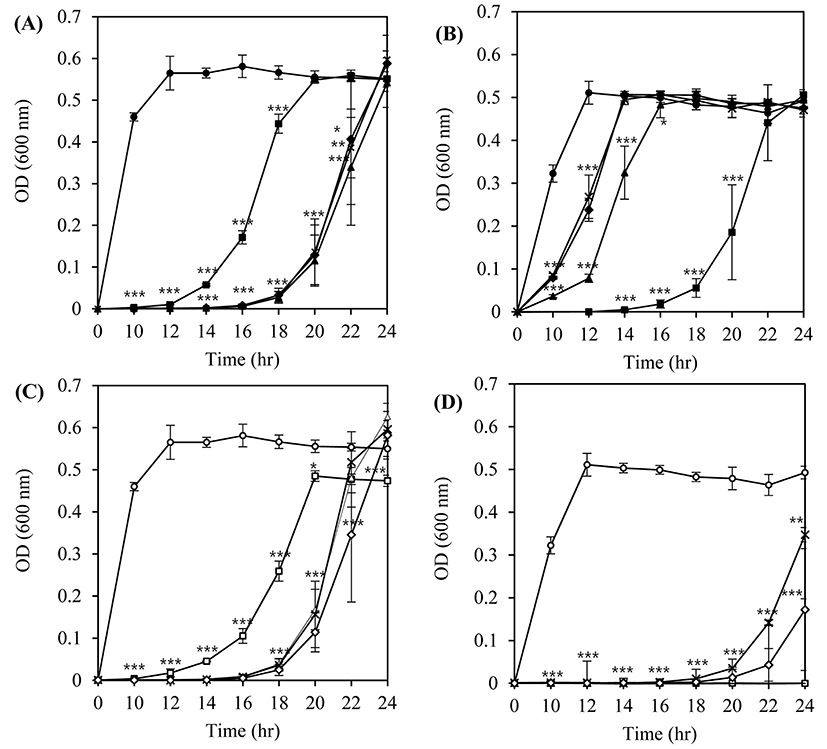
From this host-phage combination assay, we determined that use of the optimum MOI value and the phages with the highest lytic activity against a wide range of hosts or a combination of different phages (a phage cocktail) would be necessary to ensure the efficacy of bacteriophages as a biocontrol agent targeting L. monocytogenes strains with unknown serotypes or susceptibility to those phages.
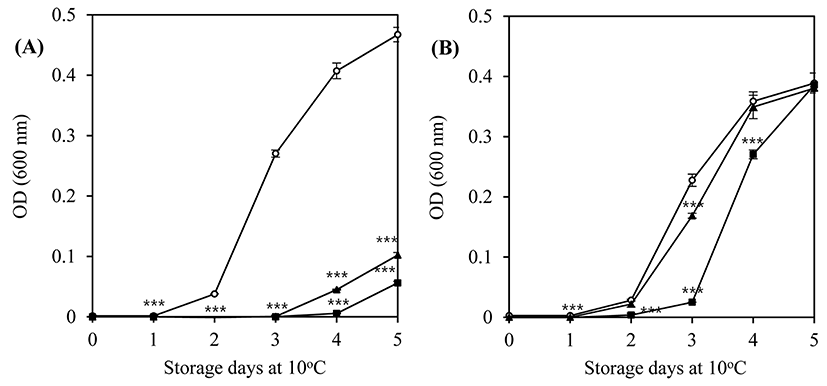
Two bacteriophages, LMP1 and LMP7, which could most effectively inhibit L. monocytogenes ATCC 19114 and L. monocytogenes ATCC 7644, respectively, were selected for further study at a lower temperature and a longer storage period. An inhibition assay in TSB at 10°C was used to investigate the potential effectiveness of bacteriophages as biocontrol agents in refrigerated food products. At an MOI of 100, LMP1 and LMP7 efficiently suppressed the growth of L. monocytogenes ATCC 7644 for up to 3 d during storage at 10°C (p<0.001) (Fig. 2A). However, neither phage was able to inhibit the growth of L. monocytogenes ATCC 19114 at an MOI of 100, though a slightly better inhibition pattern was produced by LMP1 (Fig. 2B).
The lytic activity and patterns of the phages against the host strains differed with temperature (30°C, Fig. 1 vs. 10°C, Fig. 2), which highlights the importance of temperature in the efficacy of phages in the biocontrol assay. Compared with the optimum growth temperature, low temperature can cause changes in the physiological status of L. monocytogenes, eventually influencing the cell walls, which contain receptors for the phages (Fister et al., 2016).
Electron micrographs of the bacteriophage LMP1 is shown in Fig. 3. The phage featured a long, flexible tail and an icosahedral head, which are typical morphological characteristics of the family Siphoviridae in the order Caudovirales. LMP1 consists of a head, measuring approximately 55 nm in diameter, and a long tail, ~250 nm in length (Fig. 3). This result is in accordance with those of previous reports on the morphological characteristics of these phages (Carlton et al., 2005; Dorscht et al., 2009). The listeria phages found to date are members of the order Caudovirales, which includes the family Siphoviridae as a major group (Klumpp and Loessner, 2013), and some listeria phages belonging to the family Myoviridae have also been reported (Klumpp et al., 2008). However, no phages in the Podoviridae family have yet been reported for Listeria species, which is most likely related to the structural characteristics of Listeria cells. Until now, more than 500 phages targeting the genus Listeria have been isolated and characterized to a certain extent, some of which are well-studied; studies have been done on their morphology, host specificity, and genome sequences (Klumpp and Loessner, 2013). Genome sequencing of the listeria phage LMP1 is currently underway to allow for genomic characterization of this phage and comparative genome studies with other listeria phages. Once the genome is sequenced, more systematic and another biocontrol strategy could be also developed; recombinant phage-encoded endolysins could be used to control food-borne pathogens such as L. monocytogenes (Coffey et al., 2010).

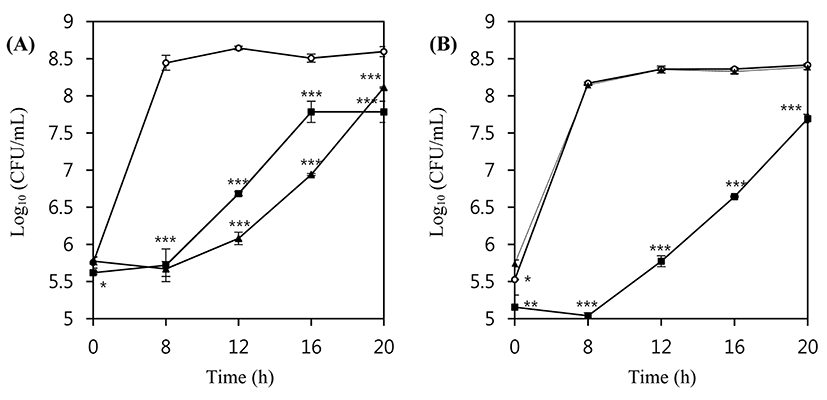
In order to examine the lytic activity of the phages in a real food matrix system, pasteurized milk from the local market was inoculated with 5×105 CFU/mL L. monocytogenes ATCC 7644 or ATCC 19114, and these suspensions were incubated at 30°C for 20 h without and with LMP1 and LMP7. Both phages efficiently inhibited the growth of L. monocytogenes ATCC 7644 at an MOI of 100 (Fig. 4A). During an 8 h incubation at 30°C, the number of L. monocytogenes ATCC 7644 cells in untreated control milk showed more than a 3 Log (CFU/mL) increase, whereas no growth or a slight decrease in the number of viable cells was observed in phage-treated host cells (p<0.001). LMP 7 showed better inhibitory activity against the growth of L. monocytogenes ATCC7644 than did LMP1, which is in accordance with the results of the liquid media assay at the same growth temperature.
In the case of L. monocytogenes ATCC 19114, LMP7 at an MOI of 100 did not show any inhibitory activity, but the addition of LMP1 produced a more than 3 log reduction compared with the untreated control (p<0.001) (Fig. 4B). The difference in the inhibitory activity of LMP7 against L. monocytogenes ATCC 19114 between tryptic soy broth and milk media is probably due to different components in the media. For example, phages in milk suspensions can be entrapped by hydrophobic and electrostatic charge interactions with some protein particles, and it has also been suggested that some phages may be inactivated by bovine whey proteins (Gill et al., 2010). Such factors potentially influence the efficacy of phage therapy by interfering with the ability of the phages to access host cells; eventually, the adsorption rate as well as reproducibility could be slowed (Fister et al., 2016).
The inhibitory activity of LMP1 and LMP7 in milk was also evaluated for 5 d at 10°C. Both LMP1 and LMP7 suppressed the growth of L. monocytogenes ATCC 7644 at an MOI of 100 (Fig. 5A). However, neither bacteriophage could suppress the growth of L. monocytogenes ATCC 19114 at an MOI of 100 (Fig. 5B).
As growth of L. monocytogenes in milk is generally slow at low temperatures, relatively long-term experiments were conducted to examine the effect of storage at 4°C for 18 d. Both LMP1 and LMP7 inhibited the growth of L. monocytogenes ATCC 7644 at an MOI of 100 (Fig. 5C), with an initial reduction at the time of application followed by further reduction at the 4 d storage time point (p<0.001). Afterward, this strain showed re-growth; however, the number of phage-treated cells did not recover up to the initial cell count during the 18 d experimental period (p<0.01). LMP1 and LMP7 were also able to reduce the initial count of L. monocytogenes ATCC 19114 right after the application of phages at an MOI of 100 (p<0.001) (Fig. 5D).
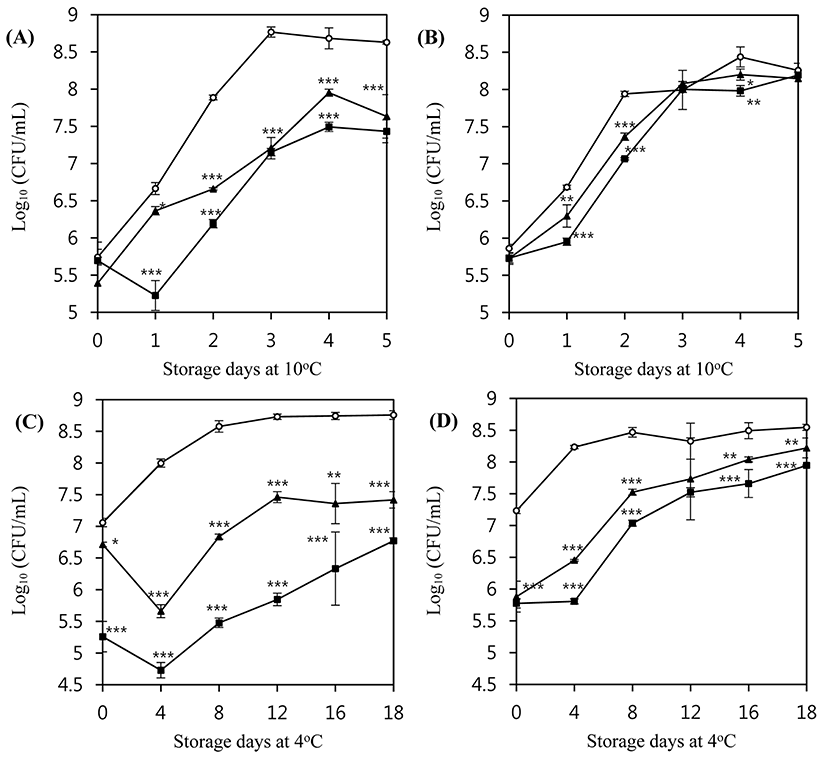
Interestingly, the inhibition pattern of L. monocytogenes by the phages in milk was different from that observed in TSB medium, especially at refrigerated temperatures. These differences may stem from the composition of the medium, and the interference would be more prominent at lower temperatures owing to increased viscosity. Some of the components of milk can act as barriers to the interaction between the bacteriophages and the host bacteria. For example, milk proteins or fat globules may interrupt a bacteriophage’s contact with the bacterial cell surface (Rodríguez-Rubio et al., 2015).
In the food industry, many bacteriophage products have been developed to control food-borne pathogens, including L. monocytogenes. For example, ListexTM P100 was developed to combat L. monocytogenes and is commercially available on the market after being designated as Generally Regarded As Safe (GRAS) by the FDA and USDA (Hagens and Loessner, 2014). Two phages found in this study, LMP1 and LMP7, are good candidates for such applications owing to their high lytic activity and wide host range as well as differences in their host specificities.
Further study is needed regarding the effectiveness of phages against L. monocytogenes under various chemical and physical conditions that are seen in a real food matrix.
Conclusions
In this study, two Listeria phages, LMP1 and LMP7, were isolated from chicken feces as a potential candidate for biocontrol of L. monocytogenes. Both bacteriophages had lytic effects on L. monocytogenes ATCC 7644, 15313, 19114, and 19115. Phages LMP1 and LMP7 were able to inhibit the growth of L. monocytogenes ATCC 7644 and 19114 in tryptic soy broth at 10°C and 30°C. In terms of host specificity, LMP1 was more effective than LMP7 against L. monocytogenes ATCC 19114. On the other hand, LMP7 was more effective than LMP1 against L. monocytogenes ATCC 7644. LMP1 showed reduced lytic activity against L. monocytogenes ATCC 7644. Based on morphological characterization, both LMP1 and LMP7 belong to the family Siphoviridae.
The lytic activity of the bacteriophages was also evaluated in milk under refrigerated conditions to confirm their potential as biocontrol agents in refrigerated foods. Both LMP1 and LMP7 were able to reduce the viable cell count of L. monocytogenes compared with that seen in untreated control milk during storage at 4°C. At an MOI value of 100, LMP1 showed better inhibition than did LMP7 against the growth of L. monocytogenes ATCC 19114 and ATCC 7644. In particular, LMP1 showed a 3 log reduction in the viable cell count of L. monocytogenes ATCC 7644.
In conclusion, two listeria phages exhibiting different host specificities were isolated and further characterized. A phage cocktail with these two phages could be a potential biocontrol agent in many food products, including dairy foods.













