Introduction
Milk is a good source of protein, with two major proteins, namely, casein and whey. Casein accounts for approximately 80% of the protein in milk (Pepe et al., 2013). Different types of casein exist, including β-casein, which comprises 25%–35% of the protein in cow’s milk. β-casein has 209 amino acids coded by the CSN2 gene and composed of 13 known variants due to differences in amino acid positions (Giribaldi et al., 2022). A1 and A2 are two variants of β-casein arising from a single nucleotide difference (A1: histidine; A2: proline) at Position 67 of β-casein (Kamiński et al., 2007). The digestion of A1 type in the small intestine of humans yields the peptide β-casomorphin-7 (BCM-7), a known μ-opioid receptor agonist that is implicated in the harmful gastrointestinal effects of milk consumption, such as affecting the digestive function (Haq et al., 2014) and causing cardiovascular and cerebrovascular diseases (McLachlan, 2001) and metabolic diseases (Ramakrishnan et al., 2020). Therefore, BCM-7 is considered a hypothetical risk factor. By contrast, the A2 variant does not generate the BCM-7 product in the small intestine of humans (Brooke-Taylor et al., 2017).
Historically, cows produced milk that contains only A2 β-casein. After natural and artificial selection, most cow’s milk that is currently available contains A1 and A2 β-casein proteins, which are obtained from A1/A2 heterozygous cows or from the milk mixture of A1A1 and A2A2 homozygous animals. In the market, A2 milk is advertised as more digestible and healthier than other types of milk. With the rapid increase in the commercial promotion and sales of A2 milk, this milk has increasingly attracted the attention of consumers and researchers. A2 milk has been noted to reduce bloating and abdominal cramps, potentially alleviating gastrointestinal symptoms linked to milk intolerance (Almuraee, 2019). However, the documented influences of A1 and A2 β-casein proteins on the gastrointestinal, endocrine, nervous, cardiovascular, and immune systems remain controversial (Fernández-Rico et al., 2022; Kay et al., 2021).
Dietary protein and its metabolites are essential nutrients for animals and humans. As one of the key elements, the intestinal microbiota produce tens of thousands of metabolites that modulate host physiology and health (Chen et al., 2019). The sources and components of dietary protein can affect gut microbiota composition and microbial metabolites (Zhao et al., 2019). At present, gut microbiota dysbiosis is involved in many diseases, including obesity, Type 2 diabetes, and inflammatory bowel diseases. The mammalian colon contains dense and diverse symbiotic microbiota, which play a particularly important role in regulating host metabolism in the colon (Bergstrom et al., 2020).
Milk with A2 β-casein variants decreased levels of the systemic inflammation marker (hs-CRP), enhanced Actinobacteria presence, and promoted better bowel habits compared to milk containing A1/A2 β-casein variants (Almuraee, 2019). Robinson et al. (2024) reported that variations in the β-casein composition of milk can impact the diversity of gut microbiota and the synthesis rates of metabolites such as aromatic amino acids and short-chain fatty acids. Animal studies have shown that A2 β-casein alleviates gut microbiota disorders in immunosuppressed mice by modulating the relative abundance of beneficial bacteria such as Oscillospira, Lactobacillus, and Bifidobacteria, while reducing levels of detrimental bacteria like Coprococcus and Desulfovibrionaceae. Moreover, A2 β-casein enhances the production of short-chain fatty acids and promotes greater diversity within the gut microbiota (Li et al., 2024). However, knowledge regarding the effects of A1 and A2 β-caseins on colon microbiota and metabolites remains limited.
The objective of the current study was to compare the effects of supplementation with A2, A2/A1, A1 bovine milk on murine body weight, intestinal morphology, intestinal cytokines, gut microbes, and serum nontarget metabolites. This study also aimed to perform an association analysis among metabolomes with microbes.
Materials and Methods
Prior to the collection of milk, a 2 mL sample of blood was drawn from each cow. Thereafter, DNA was extracted to identify the CSN2 genotype of each cow. The specific types of β-casein were determined using a proprietary method developed by our laboratory. Subsequently, A2 milk (containing A2 β-casein), A1/A2 milk (A1 and A2 β-casein), and A1 milk (A1 β-casein) was collected from the dairy farm Shandong Shijie Animal Husbandry (Dezhou, China). Raw milk was transported to the laboratory in ice packs. The milk composition of each sample was determined using Milkscan FT+/Fossmatic FC600 (FOSS Analytical A/S). The milk with the same β-casein type was grouped in accordance with primary nutrients to maintain the primary nutrients within each group to a similar extent (Table 1). After the pasteurization of fresh milk at 62°C for 30 min, the milk was frozen at –20°C for subsequent use.
| Percentage composition | A2 | A1/A2 | A1 |
|---|---|---|---|
| Protein (%) | 3.39±0.05 | 3.29±0.06 | 3.31±0.07 |
| Fat (%) | 4.41±0.09 | 4.28±0.05 | 4.44±0.02 |
| Lactose (%) | 5.28±0.15 | 5.28±0.07 | 5.29±0.03 |
Male and female C57BL/6 mice, which weighed 7–10 g and aged 3 wk, were purchased from the Pengyue Laboratory Animal Breeding (Jinan, China). The mice were housed in standard animal cages under environmentally controlled conditions of constant temperature (22°C±1°C) and relative humidity (63%±5%), with free access to water and normal chow diet (Beijing Keao Xieli, Beijing, China) during a 12 h light–dark cycle (light: 07:00–19:00). The normal chow diet composition of mice includes moisture 83 g/kg, crude protein 217.9 g/kg, crude fat 48 g/kg, crude fibre 40 g/kg, crude ash 63 g/kg, calcium 11.2 g/kg, phosphorus 8.4 g/kg. First, 32 mice were divided into 4 groups with 8 mice in each group (half male and half female). The 4 groups were supplemented with A2, A1/A2, and A1 milk, and normal saline via gavage, twice a day for 5 wk (Fig. 1). Body weight and feed intake were recorded weekly throughout the trial. At the end of the experiment, the blood, colon tissue, and their contents of all the mice were collected for metabolomics analysis, histological morphology, gene expression, and 16S ribosomal RNA (rRNA) sequencing analysis.
On the final day of the experiment, blood was collected from the orbital sinus of all the experimental mice. The blood collection method involves stabilizing the mouse by grasping the skin between its ears with the thumb and index finger of the left hand, while gently compressing the sides of the neck to restrict venous return and promote full protrusion of the eyeballs. With the right hand, hold the blood collection needle and insert its tip between the inner corner of the eye and the eyeball, gently advancing towards the throat (2–3 mm depth in mice). Stop insertion upon encountering resistance, then rotate the needle to incise the venous sinus, allowing blood to flow into the collection tube. The collected blood was then allowed to stand for 1 h at room temperature in a centrifuge tube. During that time, the blood coagulated and stratified. Subsequently, the samples were centrifuged at 1,500×g for 15 min. Thereafter, the supernatant was removed and transferred to a clean centrifuge tube. The supernatant was then aliquoted into 1.5 mL centrifuge tubes, with each tube receiving 0.2 mL. Subsequently, the samples were snap-frozen in liquid nitrogen for 15 min and subsequently stored in a −80°C refrigerator for subsequent metabolomics analysis.
Following the sacrifice of the mice via spinal dislocation, the colon was promptly excised and flushed with phosphate-buffered saline. The mid-colon was then placed in 10% neutral buffered formalin. Immediately after sacrifice, the colon tissues from each animal were snap-frozen in liquid nitrogen for subsequent analysis. Following the demise of the experimental mice, the entire intestinal tract was excised using a sterile scalpel under aseptic conditions. The contents of the colon segment were excised and then extracted with a sterile scalpel into a sterile centrifuge, which was placed on ice. The contents were divided into cryopreservation tubes and immediately frozen in liquid nitrogen. Subsequently, the samples were stored at −80°C for subsequent 16S rDNA sequencing.
The middle portion of the mouse colon was fixed in 10% formalin buffer, dehydrated with increasing amounts of alcohol, and embedded into paraffin. The paraffin-embedded samples were continuously sectioned at a thickness of 5 μm, with each section loaded into a slide and stained with hematoxylin and eosin for histopathological examination. Blind histological examination was conducted using an Olympus TH4-200 microscope (Olympus, Tokyo, Japan). The morphological indexes of the evaluation included mucosal thickness and muscular thickness. Morphometric analysis was conducted on six well-oriented and complete villi in each section of the colon. The results of this analysis was then used to describe the basic pathological changes observed in the sections. All the images were processed using ImageJ software (https://imagej.net/ij/features.html).
Total RNA was isolated from the colon tissues by using Trizol reagent (Invitrogen, Carlsbad, CA, USA), and RNA was reversely transcribed to generate complementary DNA (cDNA) by using a FastKing cDNA First-strand Synthesis Kit (Tiangen, Beijing, China). Reverse transcription quantitative real-time polymerase chain reaction (RT-qPCR) was conducted to measure the mRNA expression levels of NF-kappaB (Nf-kb), zonula occludens-1 (ZO-1), and occludin. Reactions were performed on a Roche Light Cycler 480 Real-time PCR System (Roche, Pleasanton, CA, USA). Amplification was conducted under the following conditions: 95°C for 5 min, followed by 45 cycles of 95°C for 10 s, 60°C for 10 s, and 72°C for 30 s, and followed by 95°C for 15 s, 60°C for 1 min, and 95°C for 15 s. Finally, dissociation curve analysis was performed. The specific primers of Nf-kb, ZO-1, and occludin, and the control β-actin are listed in Table 2. Gene expression was normalized to the β-actin expression by using the following formula: 2^(−ΔΔCt) method.
The genomic DNA of the colon contents was extracted using the cetrimonium bromide method. Then, the purity and concentration of DNA were detected via 1% agarose gel electrophoresis. DNA was placed into a centrifuge tube, and the sample was diluted to 1 ng/μL with sterile water.
The V3–V4 region of 16S rDNA was amplified using diluted genomic DNA as a template. 16S rRNA gene sequences were amplified using the primer pair (341F: CCTAYGGGRBGCASCAG; 806R: GGACTACNNGGGTATCTAAT) with the barcode. All PCRs were performed with 15 μL of Phusion® High-fidelity PCR Master Mix (New England Biolabs, Ipswich, MA, USA), 2 μM of forward and reverse primers, and approximately 10 ng of template DNA. Thermal cycling consisted of initial denaturation at 98°C for 1 min, followed by 30 cycles of denaturation at 98°C for 10 s, annealing at 50°C for 30 s, elongation at 72°C for 30 s, and finally, 72°C for 5 min. PCR products were purified with a Qiagen Gel Extraction Kit (Qiagen, Hilden, Germany).
Sequencing libraries were generated using a TruSeq DNA PCR-Free Sample Preparation Kit (Illumina, San Diego, CA, USA) by following the manufacturer’s recommendations, and then index codes were added. Library quality was assessed on the Qubit 2.0 Fluorometer (Thermo Fisher Scientific, Waltham, MA, USA) and Agilent Bioanalyzer 2100 system. Finally, the library was sequenced on Illumina NovaSeq 6000, and 250 bp paired-end reads were generated.
Paired-end reads were merged using FLASH (v1.2.7, http://ccb.jhu.edu/software/FLASH/). Quality filtering on the raw tags were performed under specific filtering conditions to obtain high-quality clean tags in accordance with the QIIMEv1.9.1 (http://qiime.org/scripts/split_libraries_fastq.html) quality control (QC) process.
Sequence analysis was performed using Uparse v7.0.1001 software (http://drive5.com/uparse/). Sequences with more than 97% similarity were assigned to the same operational taxonomic units (OTUs). The representative sequence for each OTU was screened for further annotation. In accordance with the principle of the algorithm, the sequences with the highest frequency in OTUs were selected as representative sequences of OTUs. Species annotation was performed on OTU sequences, and species annotation analysis was conducted using the Mothur method and the SSU rRNA database (http://ssu-rrna.org/) of SILVA138.1 (http://www.arb-silva.de/) with a threshold of 0.8–1.
Taxonomic information was obtained at each classification level. Kingdom, phylum, class, order, family, genus, and species were counted for the community composition of each sample. MUSCLE (version 3.8.31, http://www.drive5.com/muscle/) software was used for multiple sequence alignments to obtain all the OTUs on behalf of the sequence of the system. Finally, the data of each sample were homogenized, and the one with the least amount of data in the sample was adopted as the standard for homogenization. Generate stacked bar charts of species relative abundance based on the top 10 species ranked by maximum abundance in each group at the level of phylum and genus classification. Subsequent beta diversity analyses were performed on the basis of the data after homogenization. Principal coordinates analysis (PCoA) was computed based on unweighted UniFrac distances. MetaStat analysis was employed to identify differential microbial community compositions. Function prediction of these differential microorganisms was performed using Tax4Fun analysis.
First, 200 μL of serum samples were placed in an extreme pressure tube and resuspended by vortexing in prechilled 80% methanol, frozen for 5 min, and centrifuged at 15,000×g for 20 min at 4°C. A portion of the supernatant was diluted with liquid chromatography–mass spectrometry (LC–MS) grade water to a final concentration of 53% methanol. The samples were then transferred to fresh Eppendorf tubes and centrifuged at 0.9×g, 4°C for 15 min. Finally, the supernatant was injected into the liquid chromatography–tandem mass spectrometry (LC–MS/MS) system for analysis.
Ultrahigh-pressure liquid chromatography–tandem mass spectrometry (UHPLC–MS/MS; Dunn et al., 2011) was performed using a Vanquish UHPLC system (Thermo Fisher Scientific) coupled with an Orbitrap Q ExactiveTM HF mass spectrometer (Thermo Fisher Scientific) in Novogene (Beijing, China). Samples were injected onto a Hypersil GOLD column (100 mm×2.1 mm, 1.9 μm) by using a 12 min linear gradient at a flow rate of 0.2 mL/min. The eluents for the positive polarity mode were Eluent A (0.1% FA in water) and Eluent B (methanol). The eluents for the negative polarity mode were Eluent A (5 mM ammonium acetate, pH 9.0) and Eluent B. The solvent gradient was set as follows: 2% B, 1.5 min; 2%–85% B, 3 min; 85%–100% B, 10 min; 100%–2% B, 10.1 min; and 2% B, 12 min. A Q ExactiveTM HF mass spectrometer was operated in positive/negative polarity mode with a spray voltage of 3.5 kV, a capillary temperature of 320°C, a sheath gas flow rate of 35 psi, an auxiliary gas flow rate of 10 L/min, an SLens RF level of 60, and an auxiliary gas heater temperature of 350°C. QC samples that were equal-volume mixed samples of the experimental samples were used to balance the status of the LC–MS system and monitoring instruments and to evaluate the stability of the system during the entire experiment.
The raw data files generated via UHPLC–MS/MS were processed using Compound Discoverer 3.1 (Thermo Fisher Scientific) to perform peak alignment, peak picking, and quantitation for each metabolite. The major parameters were set as follows: retention time tolerance, 0.2 min; actual mass tolerance, 5 ppm; signal intensity tolerance, 30%; signal/noise ratio, 3; and minimum intensity. Thereafter, peak intensities were normalized to the total spectral intensity. The normalized data were used to predict the molecular formula based on additive ions, molecular ion peaks, and fragment ions. Then, peaks were matched with the mzCloud (https://www.mzcloud.org/), mzVault, and MassList databases to obtain the accurate qualitative and relative quantitative results. Statistical analyses were performed using the statistical software R (v3.4.3), Python (v2.7.6), and CentOS (release 6.6). When data were not normally distributed, normal transformations were attempted using the area normalization method. These metabolites were annotated using the Kyoto Encyclopedia of Genes and Genomes (KEGG) database (https://www.genome.jp/kegg/pathway.html). Principal component analysis (PCA), PCoA, and partial least squares discriminant analysis were performed at metaX (http://metax.genomics.cn). We applied univariate analysis to evaluate significance. The metabolites with variable importance in projection>1, p<0.05 and fold change (FC)≥1.2 or FC≤0.833, p<0.05 were considered distinguishingly featured metabolites.
The effects of the treatment on feed intake, body weight gain, and mRNA expression were analyzed using two-way analysis of variance (ANOVA) with Bonferroni post hoc tests. Colonic mucosal thickness and muscular layer thickness were analyzed via one-way ANOVA. All data were presented as mean±SD. All statistical analyses were performed using GraphPad Prism 9.0 (GraphPad Software, San Diego, CA, USA), and differences were considered statistically significant at p<0.05.
Results
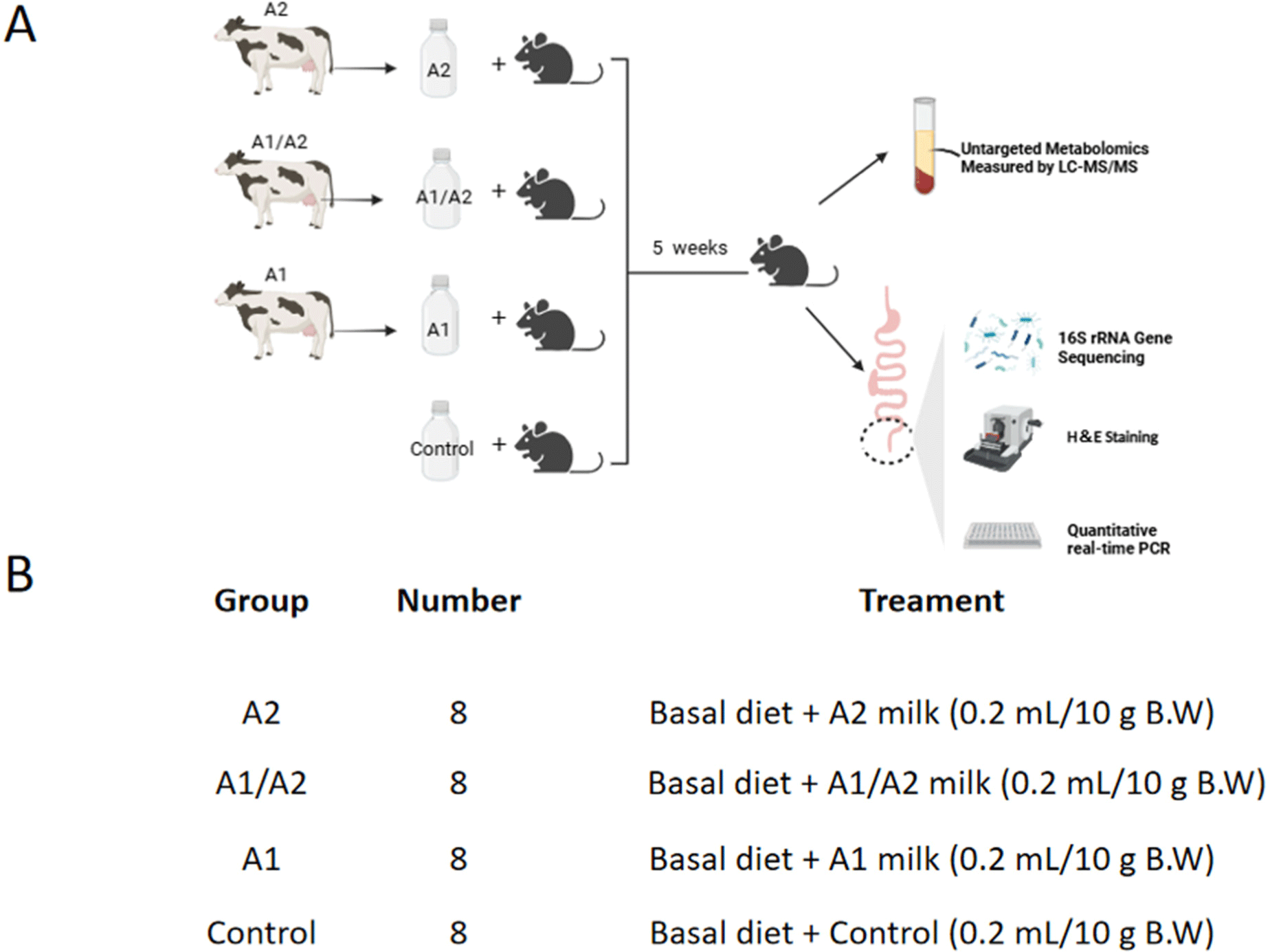
Body weight gain and feed intake were monitored every week. In the first week of experiment, the body weight gain of mice supplemented with milk with different β-caseins (A2, A1/A2, and A1) was slightly higher than that of mice given normal saline. In the second week, mice gavaged with milk that contained A2 β-casein gained more weight than mice given saline (p<0.05). In the third week, mice given A2 and A1/A2 milk gained significantly higher body weight than those given normal saline (p<0.05). In the fourth week, the body weight gain of the A1/A2 group increased significantly than that of the normal saline group (p<0.05). In the fifth week, the weight of mice in the A1/A2 group remained elevated compared with that of the saline group, although the difference was not statistically significant (p=0.0879; Fig. 2A). In addition, no significant difference was observed in the average dietary feed intake among the four groups (Fig. 2B). Body weight gain exhibited a reduction tendency in the A1 group compared with in the A2 and A1/A2 groups. These findings indicate that dietary supplementation with A2 β-casein in milk can increase body weight in mice.
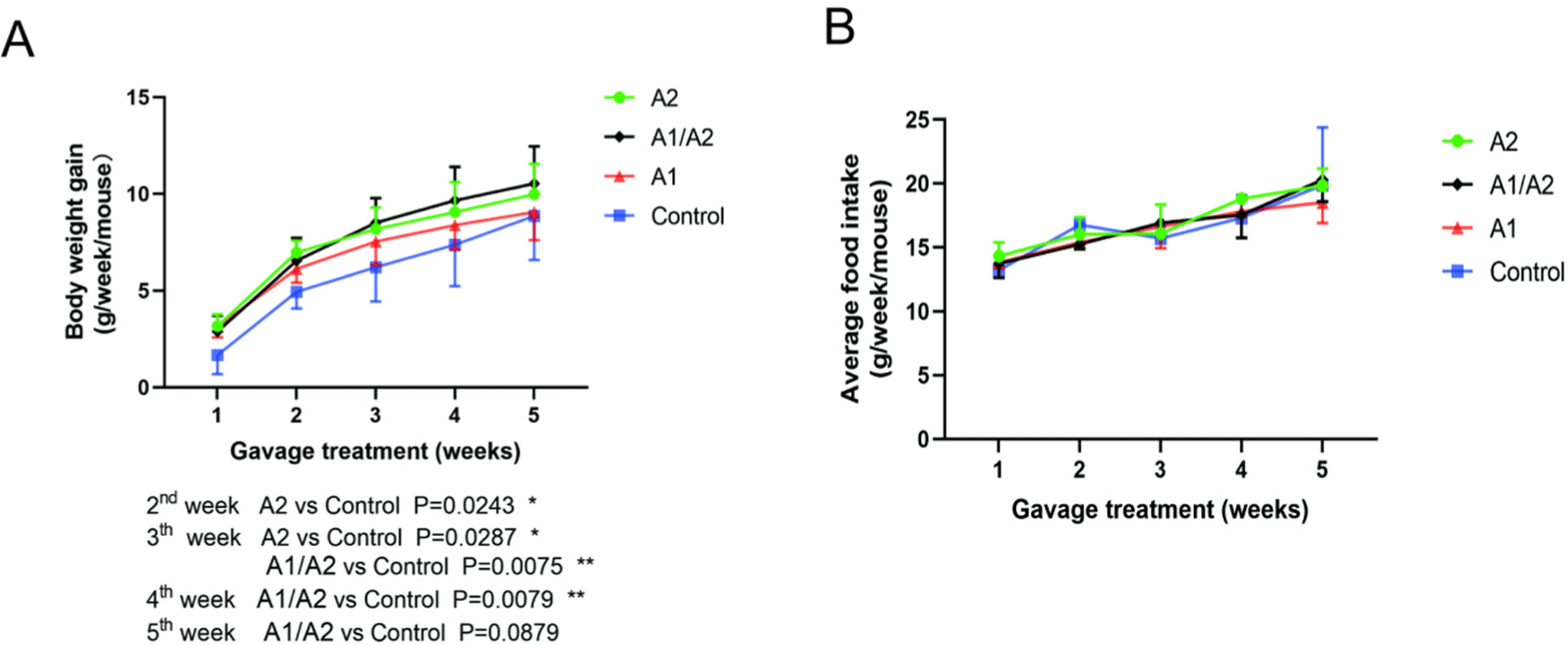
The effect of milk that contained different β-caseins on the structural integrity of the colon in mice was evaluated via morphological analysis of tissue sections. Consequently, the structure of each layer of colon tissue was generally clear in the four groups. The mucosal epithelial cells were arranged neatly in the mucosal layer. The intestinal glands in the lamina propria were closely arranged, and the shape was regular. No evident abnormality occurred in the connective tissue of the submucosa, and the muscle fibers of the muscle layer were closely arranged, without apparent inflammatory cell infiltration (Figs. 3A and B). Interestingly, the number of goblet cells increased in the A2 group, while it decreased slightly in the A1/A2, A1, and control groups (black arrows, Fig. 3B). Compared with the three other groups, the thickness of the colonic mucosa in the A2 group was higher but did not reach a significant level (p=0.07; Fig. 3C). Furthermore, the thickness of the muscle layer was similar in the four groups (Fig. 3D).
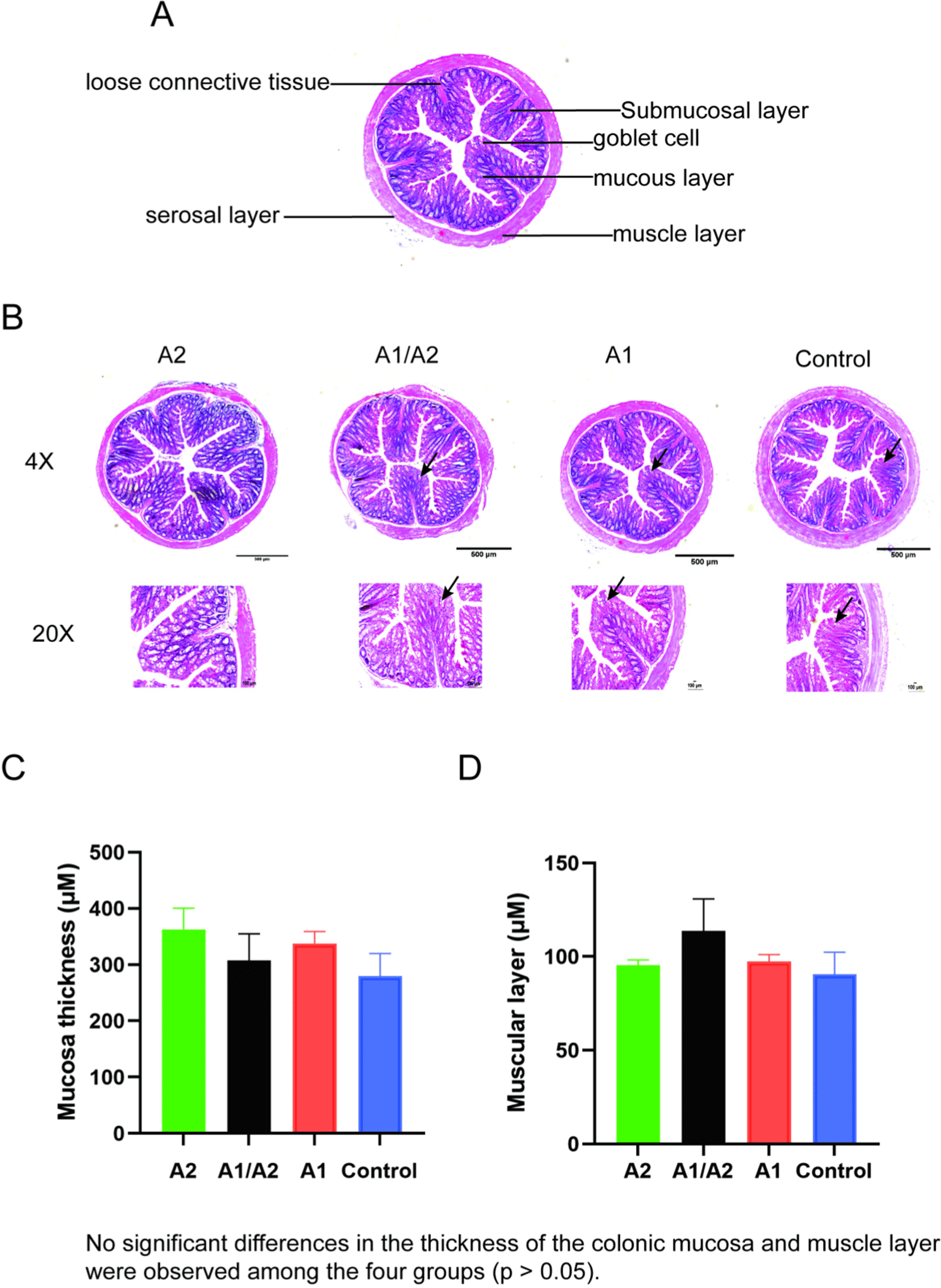
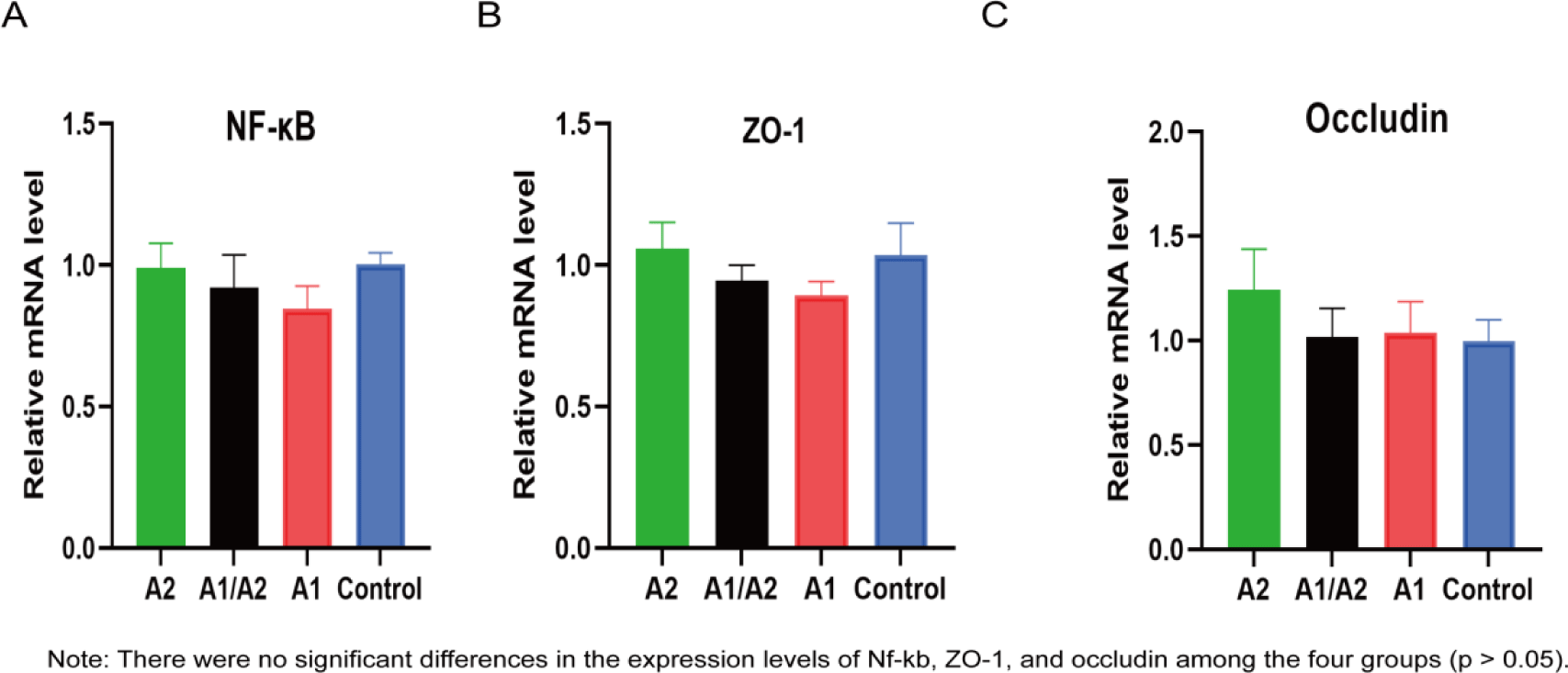
Nf-kb, a master regulator of the immune function, can suppress microbial metabolites and modulate gut inflammation (Giri et al., 2022). Tight junction proteins ZO-1 and occludin are important for the intestinal selectively permeable barrier that supports nutrient absorption and waste secretion (Kuo et al., 2022). The expression levels of Nf-kb, ZO-1, and occludin in colon tissues were investigated via RT-qPCR. No significant difference of Nf-kb, ZO-1 and occludin expression levels were observed in the four groups (Figs. 4A, B and C).
To investigate the species composition of the samples in the four groups, the effective tags of all the samples were clustered by OTUs with 97% identity, and the sequences of OTUs were annotated. As shown in Fig. 5A, 1,803, 1,549, 965, and 1,039 specific OTUs were identified in the A2, A1/A2, A1, and saline groups, respectively, along with 563 common OTUs.
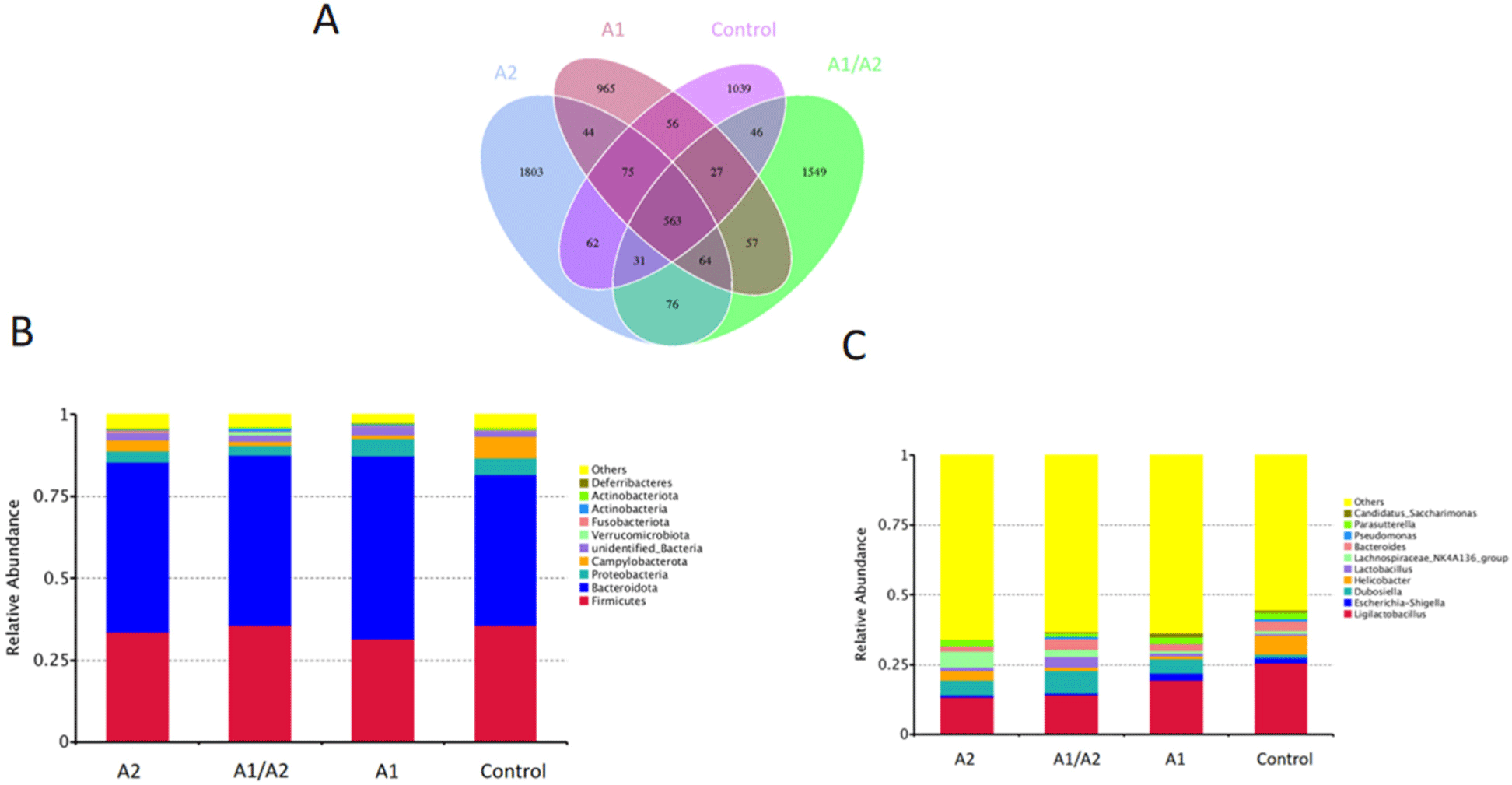
At the phylum level, the top 10 species of relative abundance were different in the four groups (Fig. 5B). Bacteroidetes and Firmicutes dominated gut microbiota in all the samples, while Campylobacterota and Proteobacteria contributed relatively smaller proportions to the intestinal microbiota community. Among them, the relative abundance of Firmicutes, Campylobacterota, Fusobacteriota, and Deferribacteres was enhanced in the A2 group. By contrast, the relative abundance of Proteobacteria was lower in the A2 group (3.4%) than in the A1 group (5.2%; Fig. 5B). At the genus level, compared with the saline group, the oral gavage of milk decreased the relative abundance of Ligilactobacillus and Helicobacter. A significant increase in the pathogenic genera Escherichia-Shigella and a decrease in Lachnospiraceae_NK4A136 was detected in the A1 group (Fig. 5C).
To further compare the microbial community composition of the samples from different groups, β-diversity was evaluated. PCoA showed that PC1 and PC2 were able to explain 16.75% and 6.39% of the variances, respectively (Fig. 6A). t-test analysis showed that at the genus level, Romboutsia (p=0.025) and Anaerostipes (p=0.049) were markedly different in the comparison between A1 and A2. In A2 versus saline, differential genera included Muribaculum, Ruminococcus, and Staphylococcus. In A1 versus saline, differential genera included Helicobacter, Muribaculum, and Anaerostipes. In A1/A2 versus saline, Helicobacter and Weissella were sharply downregulated in A1/A2, whereas Bifidobacterium, Blautia, and Coriobacteriaceae_UCG-002 were upregulated. In A2 versus A1/A2, a remarkably higher abundance of the genera Candidatus Saccharimonas and Bifidobacterium was observed in A1/A2 mice (Fig. 6B).
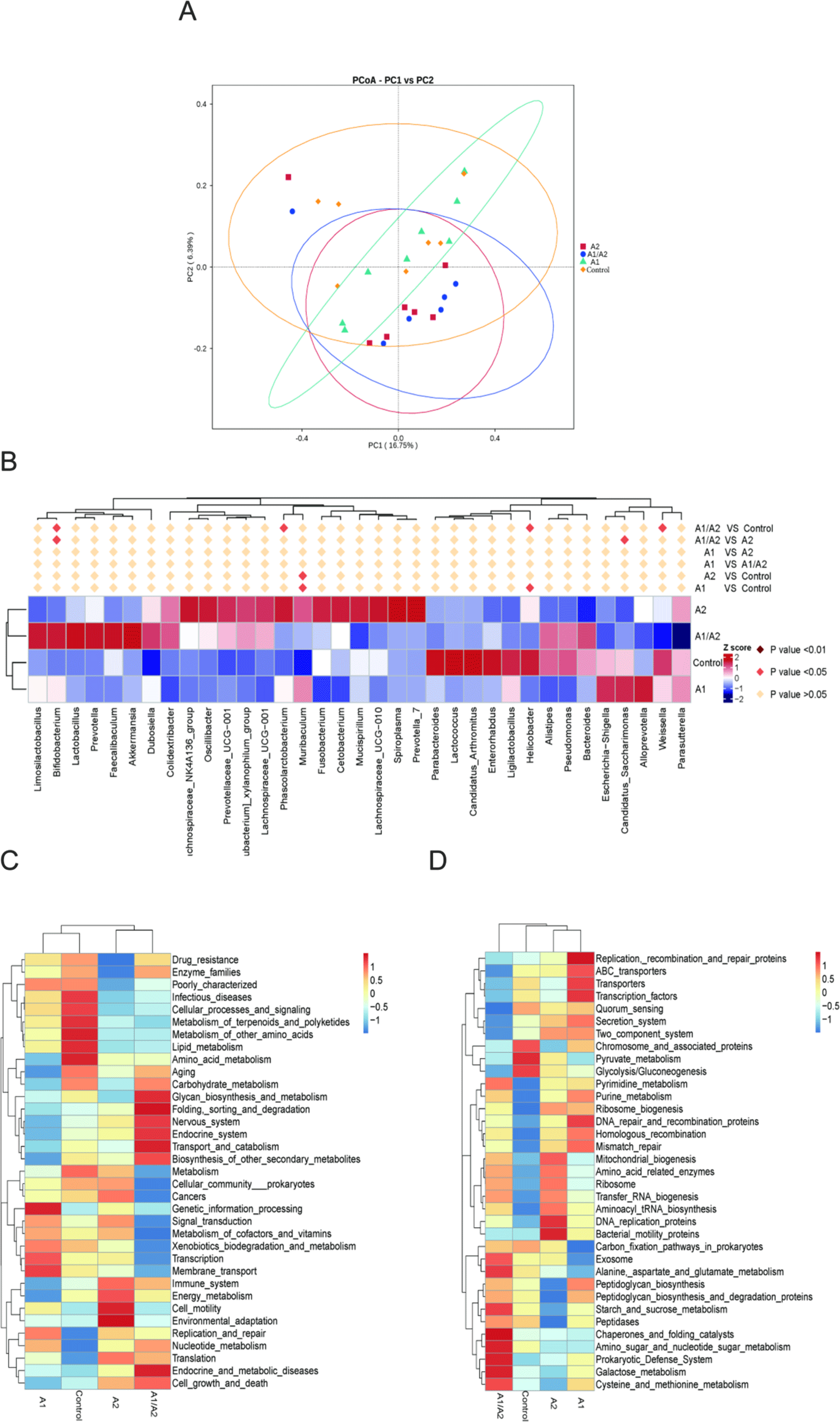
In accordance with the functional annotation and abundance information of the samples in the database, the top 35 functions in abundance and their abundance information in each sample were selected to draw heat maps. The Tax4Fun function cluster results showed that the functions of the immune system, energy metabolism, cell motility, and environmental adaptation (Fig. 6C), and the functions of bacterial motility proteins, DNA replication proteins, and mitochondrial biogenesis (Fig. 6D) were significantly enhanced in the A2 group, while the functions regarding drug resistance, infectious diseases, peptidoglycan biosynthesis, and starch and sucrose metabolism were significantly weakened. In the A1 group, significantly enhanced functions included genetic information processing, transcription, membrane transport (Fig. 6C), replication, recombination and repair proteins, ABC transporters, transporters, and transcription factors (Fig. 6D).
To evaluate the effects of milk that contained different β-caseins on serum metabolite profiles, serum samples from the four groups were collected for nontarget metabolite measurement. In PCA, small QC sample differences reflected good stability and high data quality. PC1 could account for 33.89% and 24.83% of the variations in all the samples in positive ion (Fig. 7A) and negative ion (Fig. 7B) modes, respectively. A total of 537 metabolites in positive ion mode and 371 metabolites in negative ion mode were identified in the 4 groups.
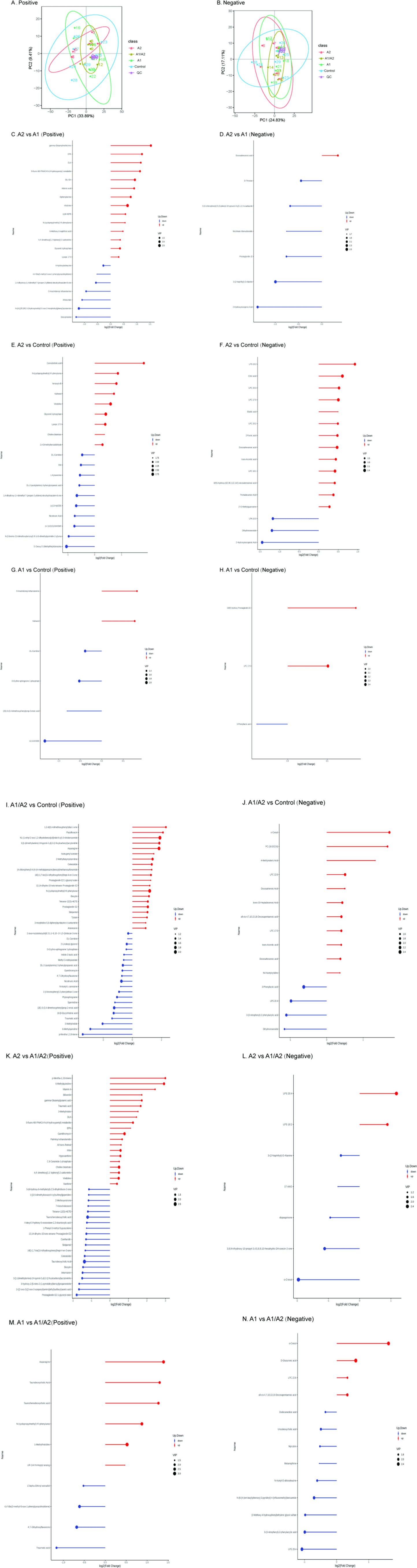
Differential metabolites of various comparisons are shown in Figs. 7B, C, D, E, F, G, H, I, J, K, L, M, and N. When the three milk-supplemented groups were compared with the control group, the A1 group had the fewest differential metabolites. Compared with the A2 and A1 groups, 14 of 21 significant differential metabolites in positive ion mode, namely, vindoline (FC=1.56, p=0.001), glycerol-3-phosphate (Gro3P; FC=1.27, p=0.004), 5-fluoro AB-PINACA N-(4-hydroxypentyl) metabolite (p=0.009), EPK (p=0.01), diphenylamine (p=0.011), Glu-Gln (FC=1.90, p=0.016), adrenic acid (FC=1.86, p=0.019), gamma-glutamylmethionine (p=0.02), lysoPC 17:0 (p=0.022), DLK (p=0.022), N-(cyclopropylmethyl)-N'-phenylurea (p=0.032), 8-HEPE (p=0.037), 6-methoxy-2-naphthoic acid (p=0.038), and 4,4'-dimethoxy[1,1'-biphenyl]-2-carbonitrile (p=0.047) were upregulated, while 7 metabolites were downregulated in the A2 group (Fig. 7C, Figs. 8A, B, C, D and E). Furthermore, 7 metabolites in negative ion mode were significantly different. Among which, 1 metabolite [docosahexanoic acid (DHA), p=0.02] was upregulated while 6 metabolites, such as 4-hydroxyisoleucine (p=0.03) and deoxyinosine (p=0.02), were downregulated in the A2 group (Fig. 7D, Figs. 8F and G). The enriched pathway terms as indicated by a comparison of the A2 and A1 groups were ferroptosis (p=0.06), purine metabolism (p=0.10), biosynthesis of unsaturated fatty acids (p=0.10), serotonergic synapse (p=0.079), and arachidonic acid metabolism (p=0.095).
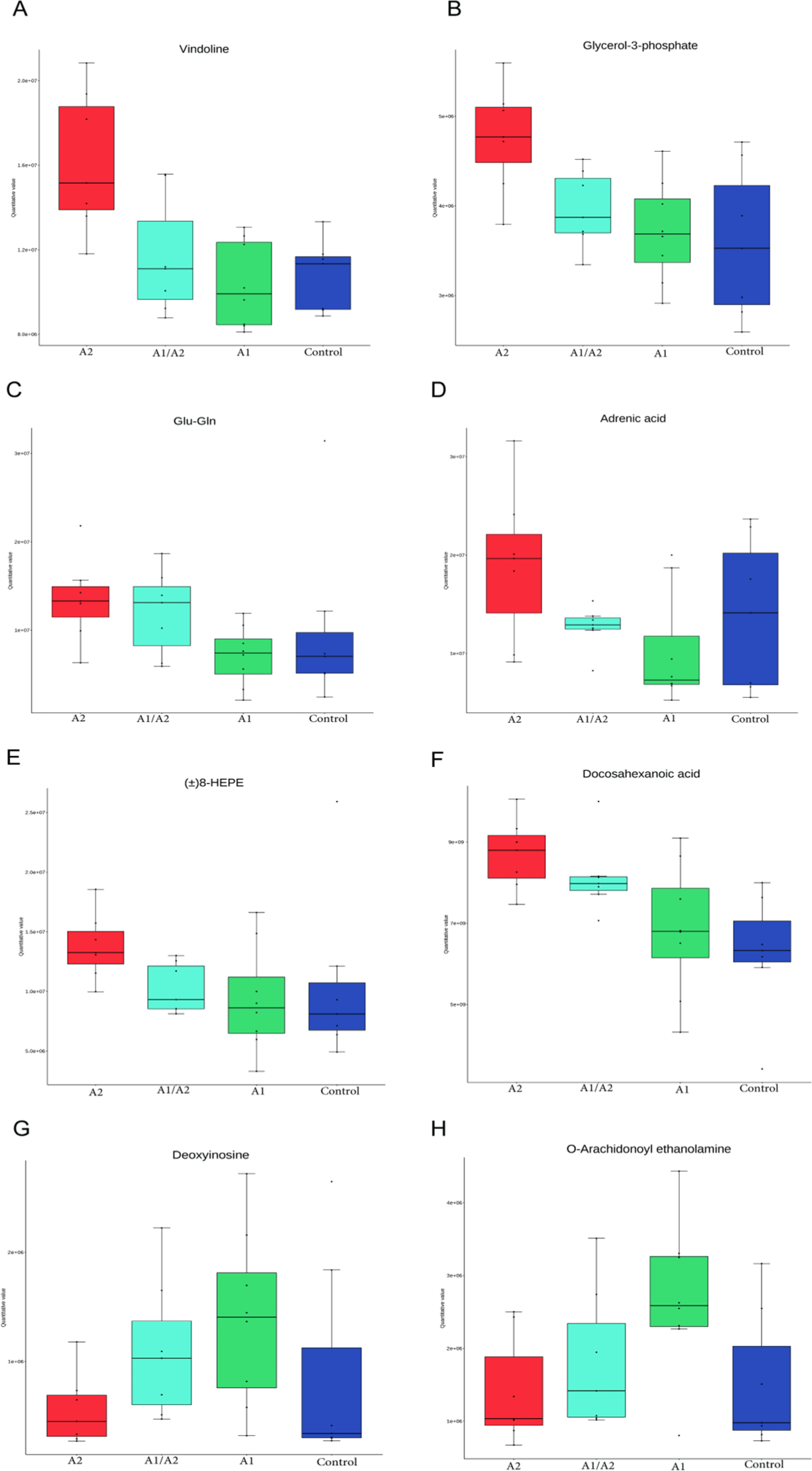
To compare with the control group, four metabolites, namely, kahweol, O-arachidonoyl ethanolamine, lysophosphatidylcholine [LPC (17:0)], and 19(R)-hydroxy prostaglandin A2, increased by 1.42–1.82 FC in the A1 group (Figs. 7G, H and Fig. 8H). LPC (17:0) was a common differential metabolite in the three comparisons (A2 versus control, A1/A2 versus control, and A1 versus control; Figs. 7E, F, G, H, I and J).
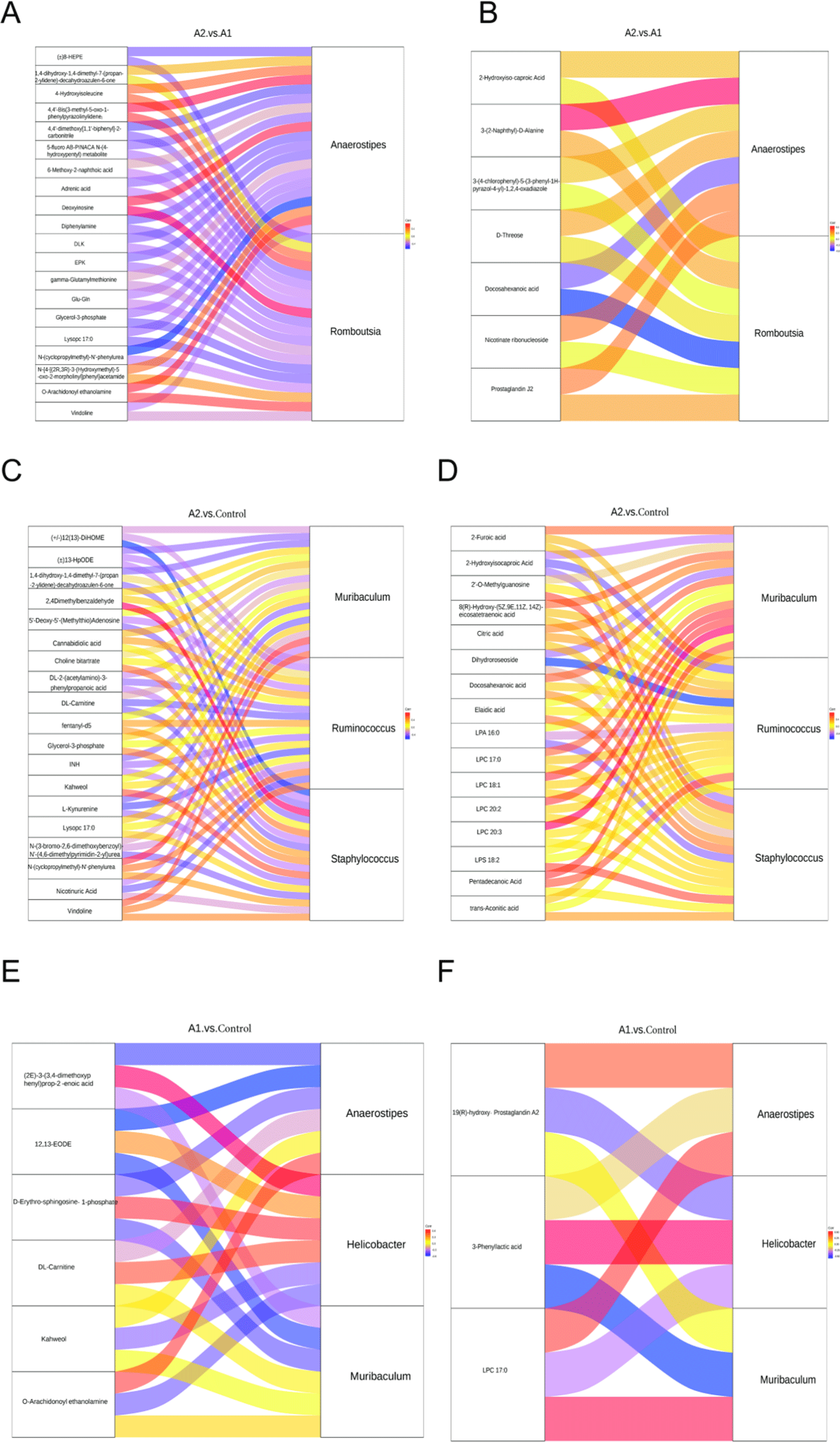
Pearson correlation analysis was conducted between bacteria genera with significant differences at the genus level obtained via 16S rDNA sequencing analysis and the differential metabolites obtained through metabolomics analysis. In the comparison of A2 and A1, the Sankey map showed that four serum metabolites (Gro3P, lysoPC 17:0, 8-HEPE, and DHA) were significantly negatively associated with Romboutsia, while three metabolites (deoxyinosine, O-arachidonoyl ethanolamine, and 4-hydroxyisoleucine) were positively associated with Romboutsia (Figs. 9A and B). In addition, negative correlations between metabolites (diphenylamine, adrenic acid, 8-HEPE, and DHA) and the genus Anaerostipes was observed. Positive correlations between metabolites (deoxyinosine, O-arachidonoyl ethanolamine, and 4-hydroxyisoleucine) and the genus Anaerostipes were found. In A2 versus saline, vindoline was positively correlated with Muribaculum. N-(cyclopropylmethyl)-N'-phenylurea was positively correlated with Ruminococcus and Muribaculum (p<0.05). Kahweol was positively correlated with Staphylococcus (p<0.05, Figs. 9C and D). In A1 versus saline, kahweol was negatively correlated with Helicobacter (p=0.08), while in A1 versus saline, kahweol was positively correlated with Staphylococcus (p<0.05, Figs. 9E and F). The correlation between distinguishing serum metabolomics and gut microbiome reflects the effect of the microbiome on metabolic activity.
Discussion
In this study, we observed changes in weight gain and colon histomorphology, and performed comprehensive analyses of related gene expression, gut microbiota, and serum metabolites by gavaging milk with different β-caseins in mice. Our findings indicated that different bovine β-caseins exerted slight influences on body weight gain and colon histomorphology while significantly altering gut microbiota and serum metabolites in mice.
In this study, the body weight gain exhibited a reduction tendency in the A1 group compared with in the A2 and A1/A2 groups. A2 β-casein is often considered to be easier on the digestive system compared to A1 β-casein because it does not release BCM-7. This could potentially lead to improved nutrient absorption and better growth outcomes in mice fed A2 milk, as fewer digestive disturbances can result in more efficient nutrient utilization.
Goblet cell is essential for the colonic mucus barrier function (Nyström et al., 2021). Goblet cells produce mucus, which serves as a crucial antimicrobial protective mechanism in the intestine. In the colon, the mucous layer produced by goblet cells acts as a barrier that inhibits direct epithelial contact with the dense microbial population. Goblet cell plays a gatekeeping role in the presentation of oral antigen to the immune system (Pelaseyed et al., 2014). In the current study, A2 milk increased the number of goblet cells in the colon. This increase suggested potential beneficial effects of A2 milk on gut health, emphasizing a specific biological pathway through which A2 milk may be advantageous. The administration of A2 milk enhanced the colonic expression of occludin, suggesting the potential role of A2 β-casein in maintaining colonic integrity and barrier function.
The discussion now includes how variations in β-casein composition may influence gut health and integrity, potentially through modulating inflammatory responses or affecting the physical properties of the gut barrier. This is crucial for understanding the dietary impacts of β-casein variants on gastrointestinal health.
Maintaining intestinal homeostasis is essential for body health. In general, the tight connections of epithelial cells are damaged in patients with digestive diseases. Occludin is a protein component of tight junctions, and the removal or reduced expression of occludin leads to loss of the gut barrier, which is important for intestinal homeostasis (Cuddihey et al., 2022; Kuo et al., 2019). A2 milk enhanced the colonic expression of occludin, suggesting the possible role of A2 β-casein in maintaining colon integrity and barrier function.
An increasing number of studies have shown that gut microbes mediate cross talk between protein metabolism and host immunity (Zhao et al., 2019). In the comparison of A1 and saline, the genus Helicobacter decreased in A1 mice. Helicobacter pylori is one of the most common infectious pathogens worldwide, causing chronic gastritis and increasing the risk of peptic ulcer disease, gastric adenocarcinoma, and mucosa-associated lymphoid tissue lymphoma (FitzGerald and Smith, 2021). A1 promoted the relative abundance of beneficial microbes, such as the genus Muribaculum (p<0.05). In the comparison of A2 and saline, two beneficial microbe genera, namely, Muribaculum and Ruminococcus, were enhanced significantly in A2 mice (p<0.05). The abundance of Muribaculaceae also differed between mice fed A1 and A2 caseins. Ruminococcus is capable of degrading cellulose and degrading and converting complex polysaccharides into a variety of nutrients for their hosts (La Reau and Suen, 2018). The genus Ruminococcus includes beneficial and harmful bacteria. For example, Ruminococcus bromii is known to have beneficial effects on health; Ruminococcus gnavus ameliorates atopic dermatitis (Ahn et al., 2022), but it is associated with Crohn’s disease, a major type of inflammatory bowel disease (Henke et al., 2019). Ruminococcus torques exacerbates the symptoms of amyotrophic lateral sclerosis, a complex neurodegenerative disorder (Blacher et al., 2019). In the comparison of A2 and A1, a reduced abundance of the genus Romboutsia was observed in A2 mice. A change in Romboutsia in the human gut is associated with genetic risk for Type 1 diabetes (Russell et al., 2019). It is one of the intestinal microbial markers in obese patients (Zeng et al., 2019).
In this study, Bacteroidetes predominated in the gut microbiota across all samples, with no significant difference observed in their abundance between A1 and A2 milk diets. However, unlike our results, Nuomin et al. (2023) reported varying levels of Bacteroidetes abundance in mice fed with A1 and A2 casein. The discrepancy can be attributed to differences in sampling materials between the two studies—one focusing on the colon and the other on the cecum. In A2 versus A1/A2, Candidatus Saccharimonas and Bifidobacterium increased in A1/A2. Candidatus Saccharimonas is the characteristic genus of female rats with autism (Gu et al., 2021). Bifidobacterium is beneficial for human physiology and pathology, most notably by modulating the host’s defense response and preventing infectious diseases (Fukuda et al., 2011). A comparison of microbial composition and function analysis did not reveal any adverse effects on health associated with A1 milk. However, our study indicates that milk protein β-caseins may influence the composition and functions of gut microbiota in mice.
Gut microbes are involved in the digestion, absorption, metabolism, and transformation of protein in the gastrointestinal tract. In the current study, several differential serum metabolites were found after gavaging milk β-caseins in mice. For example, in the comparison between the A2 and A1 groups, the upregulated metabolite in A2 mice in positive ion mode is vindoline, an indole alkaloid that exhibits hypoglycemic, antidiabetic, antioxidant, and vasodilator effects (Zhang et al., 2017). Vindoline is a natural monomer with anti-inflammatory effects. It also protects against intestinal barrier damage in mice with Crohn’s disease. Furthermore, vindoline inhibits macrophage-mediated inflammation and protects against inflammation-induced intestinal barrier damage in mice (Zhang et al., 2022). Gro3P serves as a substrate for glycerolipid synthesis and participates in the electron shuttle between cytosol and mitochondria; it is at the nexus of these pathways of glucose, lipid, and energy metabolism (Al-Mass et al., 2022). The biosynthesis of Gro3P represents an evolutionarily conserved coordinator of NADH/NAD+ redox homeostasis, contributing to the alleviation of mitochondrial disease (Liu et al., 2021). Glu-Gln plays an important role in neuronal regulation and has consistently been reported to be associated with metabolic disorders and diabetes (Wang et al., 2022). The Gln/Glu ration is significantly related to obesity development and the risk factors of some metabolic diseases (Wang et al., 2018). Adrenic acid is an ω-6 polyunsaturated fatty acid that can play a role in resolving inflammation (Brouwers et al., 2020). 8-HEPE (oxylipin) can be a promising biomarker for organic milk assessment (Samarra et al., 2021). Downregulated 4-hydroxyisoleucine is a nonprotein amino acid that inhibits obesity-related insulin resistance by reducing inflammation in mice. Downregulated deoxyinosine is a deamination product of deoxyadenosine in DNA, which is potentially mutagenic to nucleotide pairing and disrupts cellular metabolism (Kuraoka, 2015). Downregulated O-arachidonoyl ethanolamine (virodhamine, p=0.02) is an endocannabinoid that is involved in regulating intestinal permeability, fluid secretion, and immunity (Cuddihey et al., 2022).
In negative ion mode, DHA was upregulated in A2. DHA, as the dominant structural fatty acid of the brain and retina, is essential for numerous brain functions. DHA is currently considered a critical nutrient during pregnancy and breastfeeding (Comitini et al., 2020). DHA supplementation may alleviate multiple neurocognitive disease symptoms, play an important role in regulating gut microbiota (Dong et al., 2022), and attenuate tumor necrosis factor-α-induced necroptosis and autophagy (Pacheco et al., 2014). DHA is known to play a role in hematopoiesis (Kang et al., 2014) and against prostate cancer (Liang et al., 2022). Upregulated kahweol is a diterpene that exhibits anti-inflammatory, antiangiogenic, anti-oxidative, antidiabetic, and anticarcinogenic properties. Many studies have reported that kahweol plays distinct biological roles in promoting skin moisturization (Chen et al., 2021), stimulating autophagy (Seo et al., 2020), inducing apoptosis in human lung adenocarcinoma (Kim et al., 2009), ameliorating cisplatin-induced renal injury (Kim et al., 2020), and reducing food intake to lower fat (Farias-Pereira et al., 2020). The common differential metabolite in the three comparisons (A2 versus saline, A1/A2 versus saline, and A1 versus saline) was LPC (17:0). LPC (17:0) can reduce blood glucose and alleviate insulin resistance and related metabolic disorders. It is considered a potent drug candidate or health food additive for insulin resistance and hyperglycemia (Bao et al., 2022). LPC (17:0) can also reduce body weight by promoting the expression of the calcitonin receptor and inhibiting appetite (Bao et al., 2022). These metabolites participate in various physiological functions related to the health and diseases of the host.
This study involved 3-week-old male and female C57BL/6 mice administered A2, A1/A2, and A1 milk via gavage. Various factors such as age, sex, strain, and intestinal location may influence the study outcomes, which remain unexplored and require clarification in future research. Additionally, it is crucial to assess the applicability of findings from mouse models to humans when interpreting the results.
Conclusion
The supplementation of milk with different β-caseins (A2, A1/A2, and A1) has been demonstrated to enhance weight gain and gut barrier function in young mice. This effect is at least partially mediated by the regulation of gut microbiota composition and serum metabolites, which subsequently contributes to the health of the host.













