Introduction
Lactobacillus, a widely used probiotic, is recognized for its ability to extend lifespan, boost the immune system, and promote growth (Lee et al., 2022; Oh et al., 2023). Additionally, Lactobacillus species possess protective properties by preventing the invasion and colonization of pathogens and producing antipathogenic metabolites like lactic acid, hydrogen peroxide, bacteriocins, and phenylacetic acid (Chae et al., 2024; Cho et al., 2024; Eum et al., 2024; Han et al., 2024; Park et al., 2023b). They also provide immunological benefits by modulating the host’s immune function (Dimitrijevic et al., 2014; Jaafar et al., 2024). Within this group, Lactobacillus plantarum, recently denominated as Lactiplantibacillus plantarum stands out as a promising probiotic (Kim et al., 2023a; Ryu et al., 2023; Song et al., 2025; Yang et al., 2022). The safety of L. plantarum SKO-001, used in this study, has been confirmed in previous studies conducted on both mice and humans (Choi et al., 2023b; Shin et al., 2024).
Caenorhabditis elegans is an ideal surrogate animal model for studying microbe-host interactions (Kumar et al., 2020). C. elegans is extensively utilized as a model organism in research, owing to its numerous advantages, including a short lifespan, simple genetics, cost-effectiveness, ease of handling, and suitability for high-throughput screening (Choi et al., 2023a; Kang et al., 2024; Kim and Mylonakis, 2012; Lee et al., 2024b). Many research groups use C. elegans as a host model to evaluate the probiotic characteristics of various candidate strains (Kim and Mylonakis, 2012; Kim et al., 2021; Park et al., 2018). Bacteria commonly used as dietary resources for C. elegans can influence its phenotypes, including lifespan (Grompone et al., 2012; Kang et al., 2025) and immune response (Kim and Mylonakis, 2012; Park et al., 2020). Administering bacteria directly to C. elegans allows researchers to study microbe-host interactions while minimizing the influence of external nutrients. This makes C. elegans a suitable model organism for studying the characteristics of probiotics and their effects on hosts.
Whole-transcriptome analysis is commonly used to explore changes in multiple genetic pathways in C. elegans. C. elegans is well-suited for investigating genetic pathways and observing the effects of probiotics on aging and innate immunity (Kim et al., 2021; Lee et al., 2024b). Additionally, metabolomics has been used in C. elegans studies to identify metabolites linked to longevity and innate immunity (Lee et al., 2024a).
Various studies with C. elegans have shown that the consumption of specific Lactobacillus strains can enhance both lifespan and immune health (Kim et al., 2021; Lee et al., 2024a; Park et al., 2018). This study aimed to assess the probiotic properties of the candidate strain L. plantarum SKO-001 in C. elegans using multi-omics analysis.
Materials and Methods
In all experiments, the C. elegans strain fer-15; fem-1, which cannot produce progeny at 25°C, was used. Escherichia coli OP50 (OP50) was cultured in Luria-Bertani broth (BD Biosciences, Sparks, MD, USA) at 37°C for 24 h. Lacticaseibacillus rhamnosus GG (LGG) and L. plantarum SKO-001 (Accession No. KCTC 14816BP) were grown in De Man-Rogosa-Sharpe broth (BD Biosciences, Sparks, MD, USA) at 37°C for 48 h. SKO-001 was isolated from Angelica gigas Nakai and obtained from Kolmar BNH (Seoul, Korea). Four foodborne pathogenic bacteria were cultured as follows: E. coli O157:H7 EDL933 in Luria-Bertani broth at 37°C for 24 h, Salmonella Typhimurium SL1344 in nutrient broth (BD Biosciences) at 37°C for 24 h, and Staphylococcus aureus Newman and Listeria monocytogenes EGD-e in Brain Heart Infusion broth (BD Biosciences) at 37°C for 24 h.
To evaluate the colonization of SKO-001 in the C. elegans intestine, adhesion assays were conducted following established protocols (Lee et al., 2024a). C. elegans were cultured on NGM plates with OP50 until they contain eggs. Eggs were extracted using a sodium hypochlorite-sodium hydroxide solution, and synchronized L1 worms were grown on NGM plates with OP50 until the L4 stage at 25°C. Worms were then transferred to NGM plates seeded with OP50, SKO-001, or LGG [all at 8.0×109 colony-forming units (CFU/mL)]. After 48 h, 10 worms from each group were placed on Brain Heart Infusion agar with gentamycin (25 μg/mL) for 5 min. Worms were transferred to 1.5-mL Eppendorf tube containing M9 buffer with Triton X-100, then mechanically disrupted. Samples were spread on Luria-Bertani agar for OP50 and De Man-Rogosa-Sharpe agar for LGG and SKO-001, incubated at 37°C for 48 h. The experiment had 6 replicates per treatment, with 10 worms per replicate, totaling 60 worms per treatment group.
To evaluate the impact of SKO-001 on C. elegans longevity and immune response to foodborne pathogens, slight modifications were made to previously established methods (Lee et al., 2024a; Park et al., 2018).
For lifespan assay, synchronized L1 stage worms were grown on NGM agar plates with OP50 until the L4 stage at 25°C. The worms were then individually transferred to 35-mm NGM agar plates seeded with OP50, SKO-001, or LGG (all at 8.0×109 CFU/mL). Each treatment group consisted of 90 worms, split across three plates (30 worms per plate), and kept at 25°C. Survival was recorded daily, and worms were moved to fresh plates every two days. Worms were assessed as alive or dead by gently touching them with a platinum wire. The experiment continued until all C. elegans in each group died.
For killing assay, L4 stage worms were transferred to 35-mm NGM agar plates seeded with OP50, SKO-001, or LGG (all at 8.0×109 CFU/mL) for a 48 h pre-conditioning period. Worms were then moved to NGM plates containing foodborne pathogens: E. coli O157:H7 EDL933, S. Typhimurium SL1344, S. aureus Newman, and L. monocytogenes EGD-e (all at 8.0×109 CFU/mL) and kept at 25°C. Each treatment group had 90 worms, divided across three plates (30 worms per plate). Worms were enumerated daily and moved to fresh plates every two days, with survival checked by gently touching them with a platinum wire. The experiment continued until all worms in each treatment group died.
Locomotion and body dimensions were evaluated using Wormlab® software (MBF Bioscience, Williston, VT, USA), with slight modifications from the previous study (Shen et al., 2018). L4 stage C. elegans were exposed to OP50, SKO-001, or LGG (all at 8.0×109 CFU/mL) for 48 h, then moved to low-peptone NGM plates seeded with OP50. Filming began after a 10 min acclimation, with each video lasting 1 min for tracking analysis. Measurements included width, length, and peristaltic speed (μm/s). Ten worms per group were evaluated, and experiments were performed in triplicate.
The pharyngeal pumping rate, indicating food intake, was measured using a stereomicroscope by counting pharyngeal contractions over 30 s. At least 10 worms per group were measured, with all experiments conducted in triplicate.
Transcriptomic analysis was conducted with slight modifications (Ryu et al., 2021). L4 stage C. elegans were placed on NGM plates containing either OP50 (8.0×109 CFU/mL) or SKO-001 (8.0×109 CFU/mL). After a 48 h, total RNA was extracted using TRIZOL (Invitrogen, Carlsbad, CA, USA) and purified with the RNeasy Mini Kit (QIAGEN, Valencia, CA, USA). RNA-seq was performed using a TruSeq RNA Sample Prep Kit v2 (Illumina, San Diego, CA, USA) and sequenced on an Illumina NovaSeq 6000 platform with paired-end reads (2×150 bp). Trimmomatic 0.38 was used for quality trimming (Bolger et al., 2014). Reads shorter than 36 bp were discarded. Hisat2 v2.1.0 was used to create the reference genome index, and uniquely mapped reads were quantified using Subread/featureCounts v1.5.1. Genes with |Log2-fold change|>1 and p-value<0.05 were considered significantly different. In this experiment, only genes with a significant difference (p-value<0.05) and a fold change greater than 2 were included in the transcriptomic analysis. Functions of differentially expressed genes were identified using Database for Annotation, Visualization, and Integrated Discovery (DAVID), with network analysis performed using Cytoscape.
Metabolomic analysis was performed to evaluate metabolite variations using the previous method (Lee et al., 2024b). L4 stage worms were provided with either OP50 (8.0×109 CFU/mL) or SKO-001 (8.0×109 CFU/mL) for 48 h. The worms were rinsed six times with sterile deionized water, homogenized, combined with ice-cold methanol, vortexed, and centrifuged at 10,000×g for 10 min at 4°C. The supernatants were then filtered through 0.2 μm syringe filters and vacuum dried.
For the gas chromatography-mass spectrometry (GC-MS) analysis, each sample was treated with 30 μL of methoxyamine hydrochloride (Sigma-Aldrich, St. Louis, MO, USA) in pyridine (20 mg/mL) and incubated at 30°C for 90 min. Trimethylsilylation was then carried out by adding 50 μL of N,O-bis(trimethylsilyl)trifluoroacetamide (Sigma-Aldrich) and incubating at 60°C for 30 min, followed by 10 μL of fluoranthene adding (Sigma-Aldrich).
GC-MS analysis used a TRACE™ 1310 Gas Chromatograph (Thermo Fisher Scientific, Waltham, MA, USA) with an ISQ LT mass spectrometer (Thermo Fisher Scientific). Compounds were separated on a DB-5MS column (60 m×0.25 mm, 0.25-μm film thickness, Agilent, Santa Clara, CA, USA). The temperature program started at 50°C for 2 min, ramped to 180°C at 5°C/min (held for 8 min), then increased to 325°C at 2.5°C/min (held for 10 min). Samples were injected at 300°C with helium as the carrier gas at 1.5 mL/min and a split ratio of 1:60. Detection used electron ionization at 70 eV and an ion source temperature of 270°C. The mass spectrometer scanned from 30 to 450 m/z at 5 spectra/sec. Metabolites were identified using the NIST Mass Spectral Library (version 2.0, NIST, Gaithersburg, MD, USA) and data analyzed with MetaboAnalyst 5.0 (Pang et al., 2021). Only metabolites with a match score above 850 in the NIST library were included in the metabolomic analysis for this study.
The Kaplan–Meier method was used to analyze the lifespan and killing assay data for C. elegans, and results were visualized with SigmaPlot 12.0 (Systat Software, Chicago, IL, USA). Statistical analysis of other datasets was performed using Prism 9 (GraphPad Software, San Diego, CA, USA). Significance levels were set at p-values of <0.05 (*), 0.01 (**), 0.001 (***), and 0.0001 (****). Graphs are presented as mean±SEM.
Results
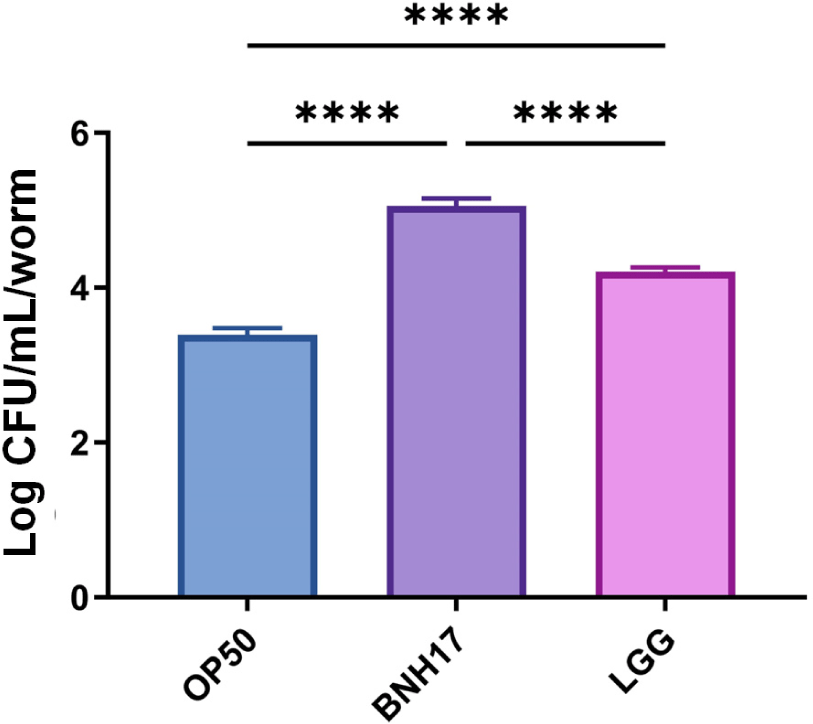
We employed C. elegans to investigate the in vivo adhesion properties of Lactiplantibacillus plantarum SKO-001 (SKO-001). E. coli OP50 (OP50) served as a negative control, while LGG was used as a positive control. The adhesion assessment was performed following 48 h of exposure to OP50, SKO-001, or LGG. Our findings revealed that SKO-001 exhibited a significantly greater adhesion capability compared to both OP50 and LGG (p<0.0001 for each comparison with OP50 and LGG; Fig. 1). This suggests that SKO-001 shows remarkable adhesive properties in C. elegans in comparison to OP50 and LGG.
We explored the impact of SKO-001 on the lifespan of C. elegans by treating the worms with OP50, SKO-001, or LGG. The group receiving OP50 was labeled as the OP50 group, the group receiving SKO-001 was labeled as the SKO-001 group, and the group receiving LGG was labeled as the LGG group. C. elegans in the SKO-001 group had a significantly longer lifespan compared to those in the OP50 group (p=0.0000; Fig. 2A). Moreover, no significant difference in lifespan was observed between the SKO-001 and LGG groups (p=0.1506; Fig. 2A). These findings suggest that SKO-001 treatment significantly prolonged the lifespan of C. elegans.
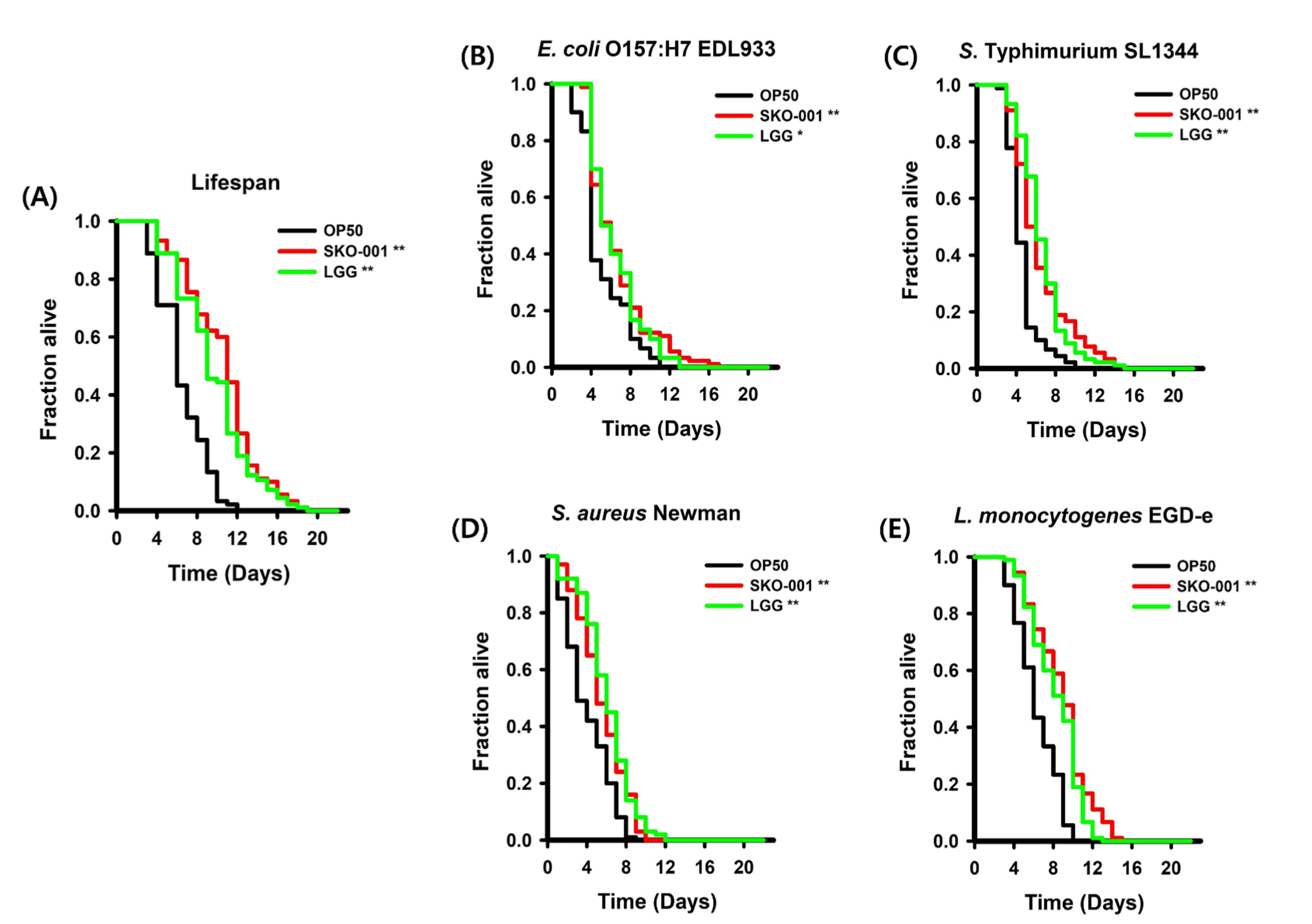
We next performed killing assays to assess whether SKO-001 improved the ability of C. elegans to defend against various foodborne pathogenic bacteria. After a 48 h pre-conditioning period with OP50, SKO-001, or LGG, the worms were placed on NGM plates seeded with foodborne pathogens. E. coli O157:H7 EDL933 and S. Typhimurium SL1344, both gram-negative bacteria, and S. aureus Newman and L. monocytogenes EGD-e, both gram-positive bacteria, were used. C. elegans that had been pre-conditioned with SKO-001 showed a significantly reduced susceptibility to the gram-negative bacteria compared to those pre-conditioned with OP50 (p=0.0003 for E. coli O157:H7 EDL933 and p=0.0000 for S. Typhimurium SL1344; Figs. 2B and C). However, no significant differences in susceptibility were observed between C. elegans pre-conditioned with SKO-001 and those pre-conditioned with LGG (p=0.6531 for E. coli O157:H7 EDL933 and p=0.8388 for S. Typhimurium SL1344; Figs. 2B and C). In experiments involving gram-positive bacteria, C. elegans pre-conditioned with SKO-001 showed significantly better survival rates compared to those pre-conditioned with OP50 (p=0.0000 for S. aureus Newman and p=0.0000 for L. monocytogenes EGD-e; Figs. 2D and E). No notable difference in survival was observed between C. elegans pre-conditioned with SKO-001 and those pre-conditioned with LGG (p=0.1506 for S. aureus Newman and p=0.3670 for L. monocytogenes EGD-e; Figs. 2D and E). Overall, these findings suggest that pre-conditioning with SKO-001 improves the resistance of C. elegans to infections caused by both gram-negative and gram-positive pathogenic bacteria.
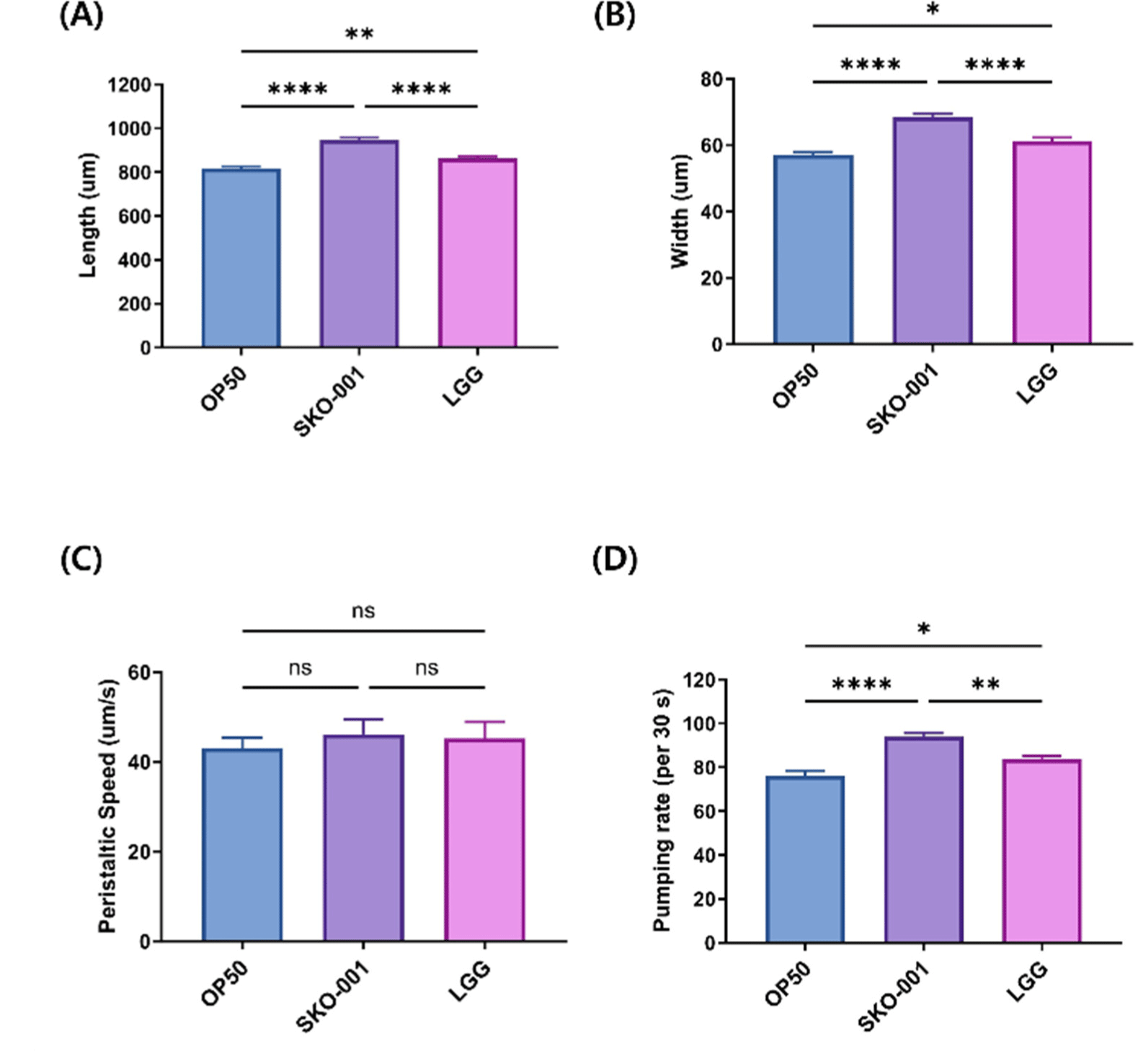
To evaluate the impact of SKO-001 on C. elegans’ phenotype, we assessed body size and locomotive activity. Worms fed SKO-001 exhibited significantly larger body dimensions, including both length and width, compared to those fed OP50 (p<0.0001 for both length and width) and LGG (p<0.0001 for both length and width; Figs. 3A and B). However, no significant difference in peristaltic speed, a measure of worm activity, was observed between the SKO-001 and OP50 groups (p=0.7777; Fig. 3C). Similarly, the peristaltic speed was comparable between the SKO-001 and LGG groups (p=0.9783; Fig. 3C). In the pumping rate assay, which reflects food intake, worms in the SKO-001 group showed a significant increase compared to those in the OP50 and LGG groups (p<0.0001 for OP50 and p=0.0015 for LGG; Fig. 3D). Overall, these findings suggest that SKO-001 enhances both body size and pumping rate in C. elegans.
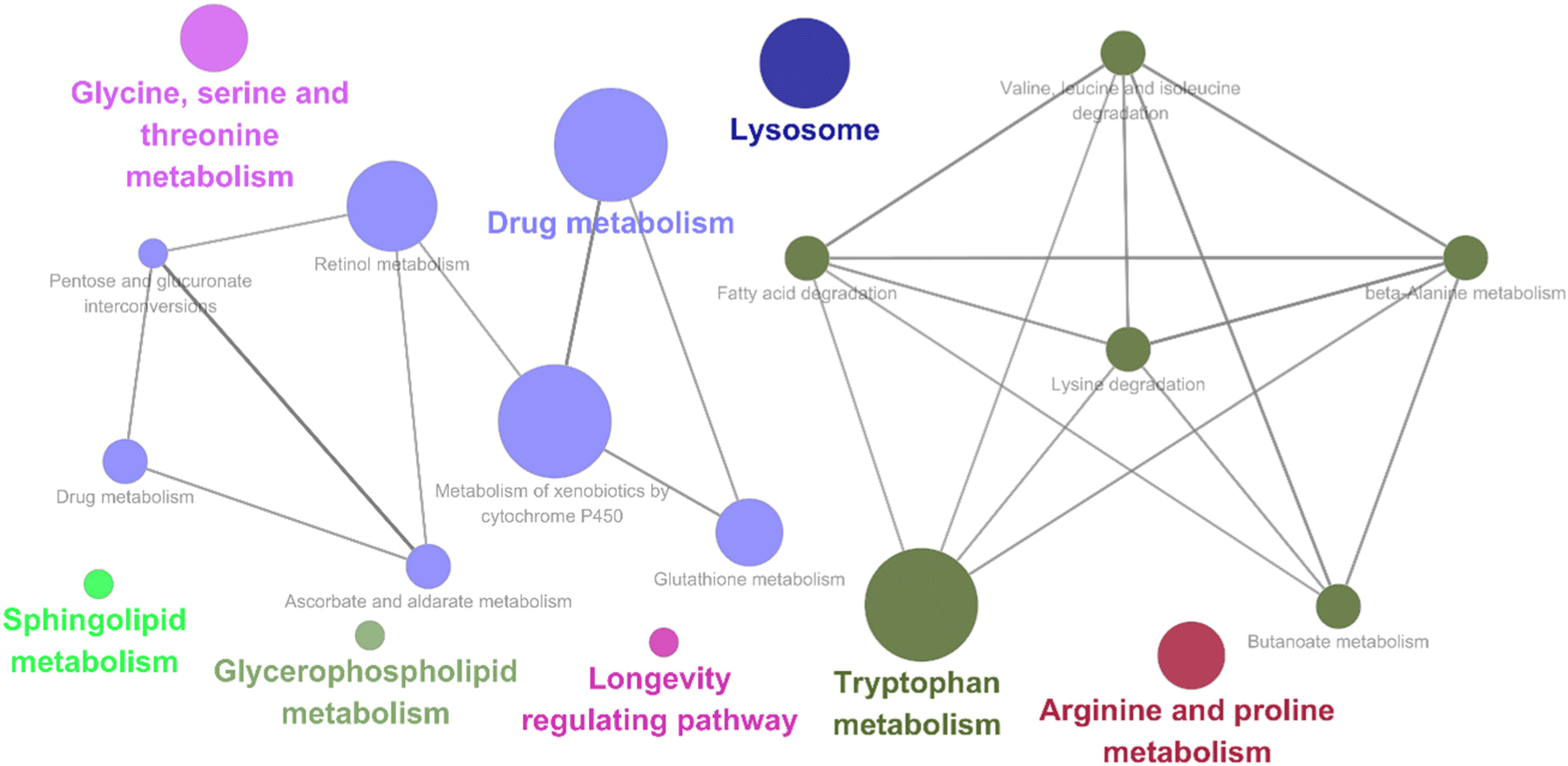
A transcriptomic analysis was conducted to investigate the gene expression alterations in C. elegans induced by SKO-001 feeding in comparison to OP50. Genes showing more than a 2-fold increase in expression with SKO-001 were identified and examined using DAVID to determine associated upregulated pathways. The top 10 pathways related to these significantly upregulated genes are detailed in Table 1. Consistent with the findings from the killing assays, pathways related to the innate immune response and defense mechanisms against both gram-positive and gram-negative bacteria were notably upregulated in SKO-001-fed C. elegans. Specifically, genes linked to C-type lectins (clec-41, clec-66, clec-86, clec-186, and clec-187) and lysozymes (lys-1, lys-2, lys-3, lys-7, and lys-8) were significantly upregulated in response to SKO-001 (Table 2). To identify the Kyoto Encyclopedia of Genes and Genomes pathways upregulated by feeding SKO-001, Cytoscape was performed with genes that exhibited more than a 2-fold increase in C. elegans fed SKO-001 compared to those fed OP50. The results identified several pathways that were significantly upregulated with SKO-001, including those involved in drug metabolism, tryptophan metabolism, lysosomes, glycine, serine, and threonine metabolism, sphingolipid metabolism, glycerophospholipid metabolism, longevity-regulating pathways, and arginine and proline metabolism (Fig. 4). These results collectively indicate that SKO-001 enhances immune response and promotes longevity.
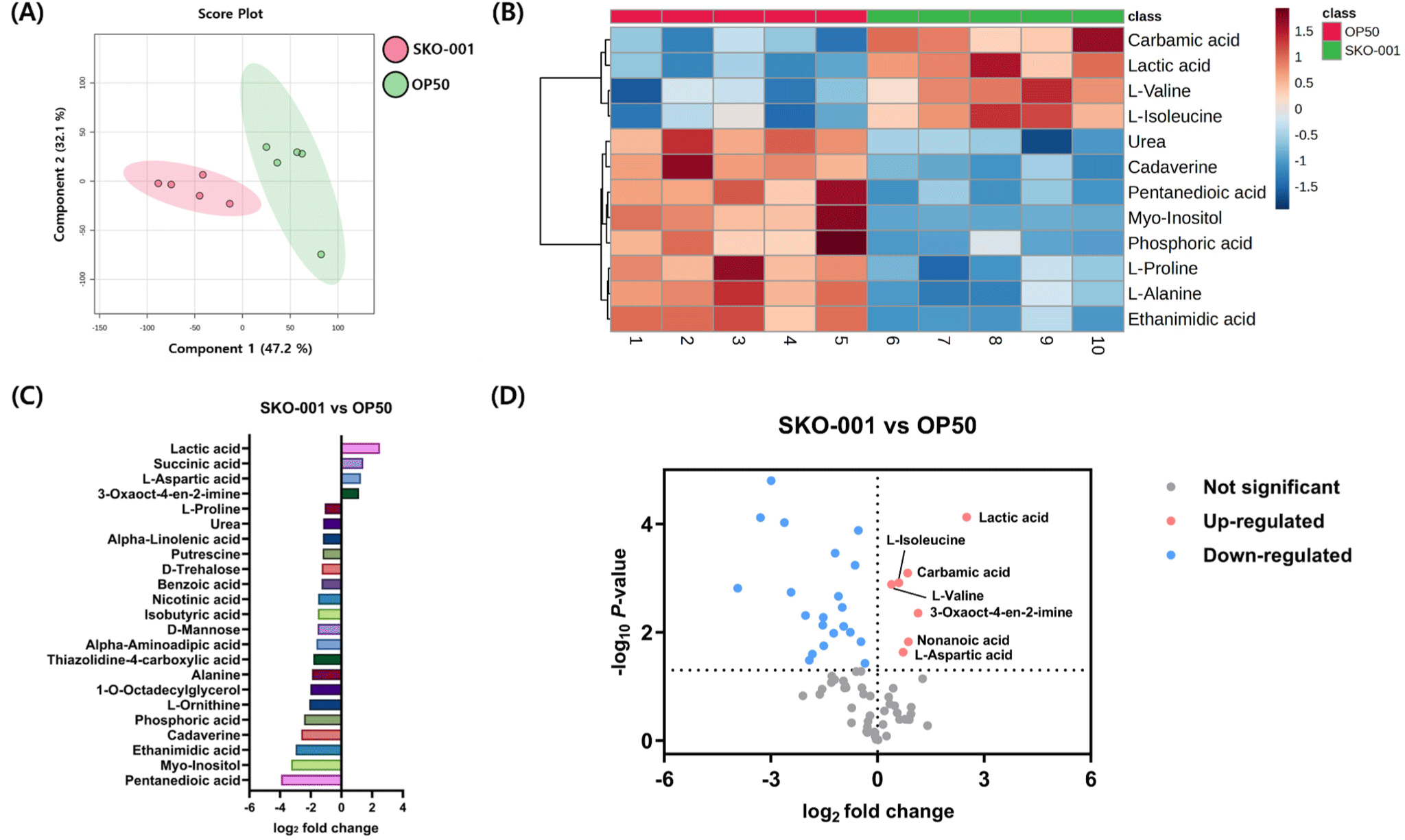
Metabolomic analysis was conducted to evaluate the effect of SKO-001 on the metabolite composition of C. elegans. The partial least squares-discriminant analysis revealed distinct clustering of metabolite profiles between C. elegans fed SKO-001 and those fed OP50 (Fig. 5A). A heatmap of the top 12 most significantly altered metabolites revealed increased levels of carbamic acid, lactic acid, L-valine, and L-isoleucine in C. elegans receiving SKO-001, compared to the OP50 group (Fig. 5B). Quantitative analysis highlighted that metabolites such as lactic acid, succinic acid, L-aspartic acid, and 3-oxaoct-4-en-2-imine were elevated by more than 2-fold in SKO-001 fed C. elegans (Fig. 5C). Additionally, a volcano plot showed that seven metabolites lactic acid, carbamic acid, L-isoleucine, L-valine, 3-oxaoct-4-en-2-imine, nonanoic acid, and L-aspartic acid were significantly upregulated in the SKO-001 group compared to the OP50 group (Fig. 5D). Overall, these findings indicate that SKO-001 alters the metabolite profile of C. elegans, with a notable increase in several key metabolites, including lactic acid.
Discussion
Lactic acid bacteria, especially those belonging to the Lactobacillus genus, are well-known for their beneficial effects on health and are frequently used as probiotics. Lactobacillus plantarum has been reported to positively influence longevity and immune responses in various studies (Kim et al., 2022; Kumar et al., 2022; Yu et al., 2022). Therefore, we investigated the potential of L. plantarum SKO-001 (SKO-001) as a probiotic candidate using C. elegans.
Previous studies have demonstrated that the ability of probiotic bacteria to adhere to the host’s gastrointestinal tract is a key criterion in their selection. This adhesive ability facilitates colonization and enhances immunomodulatory effects by stimulating the gut barrier and metabolic function (Kim et al., 2023b; Kim et al., 2024; Song et al., 2023). Consequently, probiotics can survive, proliferate, and deliver numerous health benefits to their host (Kinara et al., 2024; Oh et al., 2022; Park et al., 2023a; Park et al., 2024). In our study, SKO-001 demonstrated significantly higher adhesive ability than OP50 and LGG. Lifespan measurements are extensively used to study aging processes. C. elegans, with its short lifespan, is a suitable in vivo model for measuring the ability of candidate probiotic bacteria (Tissenbaum, 2015). In the lifespan assay, SKO-001 significantly increased the lifespan of C. elegans compared to OP50, showing no significant difference from LGG. This result supports earlier findings that Lactobacillus species with probiotic properties can increase the lifespan of C. elegans (Heo et al., 2018; Lee et al., 2024a). Similarly, the killing assay revealed that pre-conditioning with SKO-001 significantly improved the immune response of C. elegans against both gram-negative and gram-positive pathogenic bacteria. This observation aligns with previous research, which underscores the strong antimicrobial properties of L. plantarum against pathogens and its capacity to boost the immune response in C. elegans (Li et al., 2017; Mun et al., 2019).
The quality of food affects worm phenotypes (Shtonda and Avery, 2006). Additionally, different bacteria can impact the growth of C. elegans to varying degrees (Avery and You, 2018). Therefore, we measured worm size and locomotor activity to assess the quality of SKO-001 as a food source and to determine whether it could alter the phenotype of the worms. Worms fed SKO-001 exhibited a significant increase in both length and width compared to those fed OP50 and LGG. Furthermore, SKO-001 also improved the pumping rate more effectively than OP50 and LGG. These results indicate that SKO-001 not only caused notable phenotypic changes in the worms but also enhanced their growth performance.
Our study indicates that pre-conditioning C. elegans with SKO-001 enhances its immune defense against foodborne pathogens. We hypothesized that pre-conditioning upregulates specific immune-related genes. Transcriptomic analysis revealed that genes with more than a 2-fold increase in C. elegans fed SKO-001, compared to those fed OP50, were predominantly associated with innate immunity.
Notably, genes related to C-type lectins (clec-41, clec-66, clec-86, clec-186, and clec-187) and lysozymes (lys-1, lys-2, lys-3, lys-7, and lys-8) showed significant upregulation following SKO-001 treatment. In C. elegans, clec genes encode a variety of proteins with C-type lectin-like domains, which play a role in pathogen defense (Schulenburg et al., 2008). Previous studies have shown that clec-41 plays a vital role in the resistance to the gram-positive pathogen Bacillus thuringiensis MYBt18247 (Pees et al., 2021). Similarly, clec-86 has been demonstrated to be essential for defense against the gram-positive pathogen Microbacterium nematophilum (O’Rourke et al., 2006). These results suggested that clec-41 and clec-86 play a crucial role in resistance against pathogenic bacteria. Consistent with prior studies, the expression levels of both clec-41 and clec-86 were notably higher in C. elegans fed SKO-001 compared to those fed OP50. Lysozymes function as antimicrobial agents within the C. elegans gut, breaking down bacterial cells (Ciancio, 2016). Genes related to lysozymes, including lys-1, lys-3, and lys-7, are essential for the defense mechanisms in C. elegans (Schulenburg et al., 2008). We found that these genes were significantly upregulated following SKO-001 feeding. This suggests that the enhanced expression of both C-type lectin and lysozyme-related genes induced by SKO-001 contributes to a more robust immune response against pathogenic bacteria.
In the metabolomic analysis, C. elegans fed SKO-001 showed a notable increase in lactic acid compared to those fed OP50. Lactic acid, often produced by lactic acid bacteria, is associated with enhanced defense and resistance in C. elegans (Fernández et al., 2003). The higher levels of lactic acid observed with SKO-001 treatment likely contributed to an improved immune response against pathogens. Additionally, the branched-chain amino acids L-isoleucine and L-valine, which were elevated in SKO-001 fed C. elegans, are crucial for various physiological processes. Previous studies have demonstrated that supplementing C. elegans with L-valine and L-isoleucine can significantly prolong their lifespan (Wang and Zhang, 2018; Wang et al., 2018). Collectively, the increased levels of metabolites observed with SKO-001 feeding may have contributed to enhanced longevity and improved immune response in C. elegans.
Conclusion
In conclusion, we investigated the probiotic potential of L. plantarum SKO-001 (SKO-001) using C. elegans as a model organism. SKO-001 showed superior adhesion capabilities compared to OP50 and LGG, suggesting its effectiveness in gastrointestinal colonization. Additionally, SKO-001 significantly prolonged the lifespan of C. elegans, improved its resistance to foodborne pathogens, and supported its growth. Transcriptomic analysis revealed notable upregulation of genes related to the innate immune system, particularly those involved in C-type lectins and lysozymes. Metabolomic analysis showed increased levels of lactic acid, L-valine, and L-isoleucine in C. elegans treated with SKO-001. Overall, our findings suggest that L. plantarum SKO-001 is a promising probiotic with potential benefits for improving longevity and boosting immune function.













