Introduction
Meat, which constitutes approximately one fourth of the protein intake in the human diet and provides 15% of the energy consumed, plays a vital role in the growth and maintenance of the human body (Alexander et al., 2017). Furthermore, owing to the continuous increase in the global population, meat consumption is steadily increasing, with an estimated 2.55 million tons expected by 2050 (Organisation for Economic Co-operation and Development [OECD] and Food and Agriculture Organization of the United Nations [FAO], 2021). This escalating demand for meat production and consumption raises concerns about environmental issues and resource depletion, highlighting the need for sustainable meat production methods (Parlasca and Qaim, 2022). Issues related to the environment, animal welfare, and health have led to a rapid expansion of research on meat analogue (Green et al., 2022). Among these meat analogue, cell-based foods, edible insects, and plant-based meats are gaining popularity.
Cell-based food is currently recognized by the FAO as a standardized term (FAO, 2023). It is receiving attention as a new technology that can meet the increasing demand for meat, while overcoming the limitations of traditional meat production methods. Livestock farming, especially cattle rearing, has a significant impact on the environment, health, and animal welfare (Ramani et al., 2021). In this context, cell-based food produced through in vitro cell cultivation has the potential to serve as a safe and efficient alternative to traditional slaughter-based meat production methods. Muscle is a critical tissue responsible for animal movement and body structure, and the development and differentiation of muscle cells play a pivotal role in muscle tissue formation (Mukund and Subramaniam, 2020).
The cells used in cell-based meat research are skeletal muscle satellite cell (SMSCs) located between the sarcolemma and basal lamina (Ding et al., 2017). These SMSCs are essential for muscle growth, repair, and regeneration and can influence changes in muscle conditions (Kim et al., 2022). The myogenic differentiation ability of SMSC depends primarily on the expression of Pax3 and 7 genes and muscle regulatory factors (MRFs), including MyoD, Myf5, Myogenin, and MRF4 (Asfour et al., 2018). Sequential activation and inhibition of Pax3/7 and MRFs are essential for muscle-forming processes in muscle cells (Collins et al., 2009). Pax7 is expressed in all SMSC and is indispensable for postnatal maintenance and self-renewal (Seale et al., 2000). Myogenin regulates the development and differentiation of muscle cells, and is expressed during the formation of muscle tissue (Zhang et al., 2020). MRF4, also known as MYF6, is involved in the development of muscle cells, and regulates their differentiation and growth (Shirakawa et al., 2022). MHC1 is a protein related to muscle tissue contraction and is an important gene expressed when muscle cells differentiate, mature, and form muscle tissue (Kim et al., 2023).
Hanwoo is divided into steers and cows, and differences exist between their meat characteristics including taste (Cho et al., 2020). Steers are known to have relatively mild-tasting meat, whereas cow meat has a rich, salty flavor (Gajaweera et al., 2020; Joo et al., 2017). Various factors, such as age, and sex, may lead to differences in the growth rate and genetic makeup of SMSC in Hanwoo cattle (Kim et al., 2023). Previous studies mainly conducted sex-based comparative analyses of human and mouse SMSC. In addition, a previous study comparing males (bulls and steers) and females (heifers) showed that bulls had faster growth than heifers (Reyneke, 1976). Despite this finding, there is still a lack of sex-based comparative analyses of SMSC. Therefore, this study aimed to investigate the differences in muscle cell growth and differentiation between Hanwoo steers and cows.
Materials and Methods
Dulbecco’s Modified Eagle Medium-F12 (DMEM-F12), fetal bovine serum (FBS), horse serum (HS), and antibiotic-antimycotics (AA) from Gibco (Thermo Fisher Scientific, Waltham, MA, USA) and Dulbecco’s phosphate-buffered saline (DPBS) were obtained from Welgene (Gyeongsan, Korea). For RNA extraction, cDNA synthesis, and quantitative real-time polymerase chain reaction (PCR), Trizol reagent (AccuzolTm Total RNA Extraction Reagent, Bioneer, Seoul, Korea), diethylpyrocarbonate (DEPC) water (Bioneer), a cDNA transcription kit (AccuPower CycleScript RT PreMix, Bioneer), quantitative PCR (qPCR) MasterMix (Bioneer), and nuclease-free water (Ambion®, Austin, TX, USA) were used.
Thirty three months of bovine muscle tissues (three steers and three cows) were purchased from Farmstory Hannaeng Bio & Food (Cheongju, Korea). Animal sampling methods were used for ethical approval. Muscles were obtained from the top round muscle and transport to the lab. After that, tissues were rinsed with 70% ethanol and washed three times with DPBS. The tissues were minced using a meat grinder and digested with 0.8 mg/mL Pronase (Sigma-Aldrich, St. Louis, MO, USA) for 40 min at 37°C with vortexing every 10 min. After incubation, the tubes were centrifuged at 1,200×g for 15 min and the pellets were resuspended in DMEM-F12 containing 10% FBS. The supernatant was discarded, and DMEM-F12 containing 10% FBS was added to the tubes, mixed and centrifuged at 300×g for 5 min. The supernatant was filtered through a 100 μm cell strainer, collected in 50 mL tubes and centrifuged at 1,200×g for 15 min. The final step was collection in GM containing 10% dimethyl sulfoxide until used. Isolated bovine SMSC were cultured in a T-25 flask containing DMEM-F12, 10% FBS, and 1% AA. In passage 3, muscle satellite cells were isolated using Magnetic-Activated Cell Sorting with CD29, a marker known for muscle cells. The experiment was conducted using the same method described in the previous study (Kim et al., 2023).
The steers and cows of the Hanwoo SMSC were seeded at 2×104 cells/mL in a 6-well plate. SMSC were cultured in growth medium [DMEM-F12+GlutaMax (Gibco, Gaithersburg, MD, USA)+1% AA+10% FBS]. After 24 h, the growth medium was replaced with fresh medium, and the cell number was determined. The medium was replaced with fresh medium every 48 h. Trypsinization for cell counting was performed using 0.05% trypsin-EDTA (Gibco) after 1, 3, 5, and 7 days. Cell number was analyzed using a hemocytometer (Counting Chamber; Paul Marienfeld, Wöllerspfad, Germany). Measurements for each sample were taken in triplicate and averaged daily.
The steers and cows of the Hanwoo SMSC were seeded using 4×104 cells/mL in a 6-well plate in order to check the myogenic cell differentiation. SMSC were cultured in a growth medium consisting of DMEM-F12/GlutaMax containing 1% AA (Gibco) and 10% FBS (GM; Gibco). When the confluency of the cells reached 80%, the medium was changed to a medium containing DMEM-F12, 1% AA, and 2% HS (DM; Gibco) and was replaced every 24 h. When SMSC started to differentiate, their morphology was analyzed. Cell differentiation was evaluated at 24, 48, 72, and 96 h. For cell differentiation analysis, SMSC were stained using May–Grünwald solution (Sigma-Aldrich) and Giemsa stain solution (Sigma-Aldrich). After removing the medium, the cells were washed twice with DPBS, fixed with 100% methanol for 10 min, and methanol was subsequently removed. May-Grünwald solution was added for 5 min, followed by dilution with distilled water. After removing the solution, Giemsa staining solution (1:20) was then added for 20 min. Finally, the solution was removed, and the stained cells were examined under a microscope (CKX53, Olympus, Tokyo, Japan).
Total RNA was isolated using TRIzol reagent (AccuzolTM Total RNA Extraction Reagent, Bioneer) according to the manufacturer’s guidelines and previous protocol (Kim et al., 2023). Next, 20 μL of DEPC water (Bioneer) was added, and the purity and concentration of RNA were measured using a Microplate Spectrophotometer (Multiskan Sky, Thermo Fisher Scientific) and a μDrop plate (μDropTM, Thermo Fisher Scientific). The total RNA concentration was determined to adjust to 1 μg of RNA. The RNA samples were then reverse-transcribed into cDNA using a cDNA reverse transcription kit (AccuPower CycleScript RT PreMix; Bioneer) with a GeneAmp PCR System 9700 (Applied Biosystems, Singapore). The machine was run based on the cDNA synthesis condition, which consisted of the synthesis step at 45°C for 60 min and the heat inactivation step at 95°C for 5 min.
qPCR was performed on the cDNA using AccuPower® 2X Greenstar qPCR MasterMix (Bioneer) and a StepOnePlus Real-Time PCR system (Applied Biosystems, Singapore) following the manufacturer’s guidelines and a previous study (Kim et al., 2023). All the samples were recorded in triplicate, and the primer sequences for housekeeping and target genes, as well as the cycling temperatures, are provided in Tables 1 and 2. And the data was used in order to analyze it using the ΔΔCt method.
| Gene | Primer sequence (5’–3’) | Reference |
|---|---|---|
| GAPDH | F: ACTCTGGCAAAGTGGATGTTGTC | Li et al. (2019) |
| R: GCATCACCCCACTTGATGTTG | ||
| PAX7 | F: TGCCCTCAGTGAGTTCGATT | Li et al. (2019) |
| R: CGGGTTCTGACTCCACATCT | ||
| MyoG | F: AGAAGGTGAATGAAGCCTTCGA | de Las Heras-Saldana et al. (2019) |
| R: GCAGGCGCTCTATGTACTGGAT | ||
| MRF4 | F: GGTGGACCCCTTCAGCTACAG | Shibata et al. (2006) |
| R: TGCTTGTCCCTCCTTCCTTGG | ||
| MHC1 | F: CCCACTTCTCCCTGATCCACTAC | Ahn et al. (2020) |
| R: TTGAGCGGGTCTTTGTTTTTCT |
Calculation of ΔΔCt was conducted as follows. First, the cycle threshold was obtained for each sample by qPCR. Second, in order to calculate the ΔCt values, the Ct value of the housekeeping gene was subtracted from that of the target gene. Third, the ΔΔCt values were obtained by subtracting the ΔCt value of the steer group as the control from the ΔCt value of the cow group. Finally, the relative quantity value was obtained by calculation of 2–ΔΔCt.
All data were collected through experiments conducted in triplicate, and the results are presented as the mean±SD. Statistical analysis was performed using two-way analysis of variance in Prism 9.4.0 (GraphPad Software, San Diego, CA, USA), and statistical significance was defined at a significance level of * p<0.05, ** p<0.01, *** p<0.001, and **** p<0.0001.
Results and Discussion
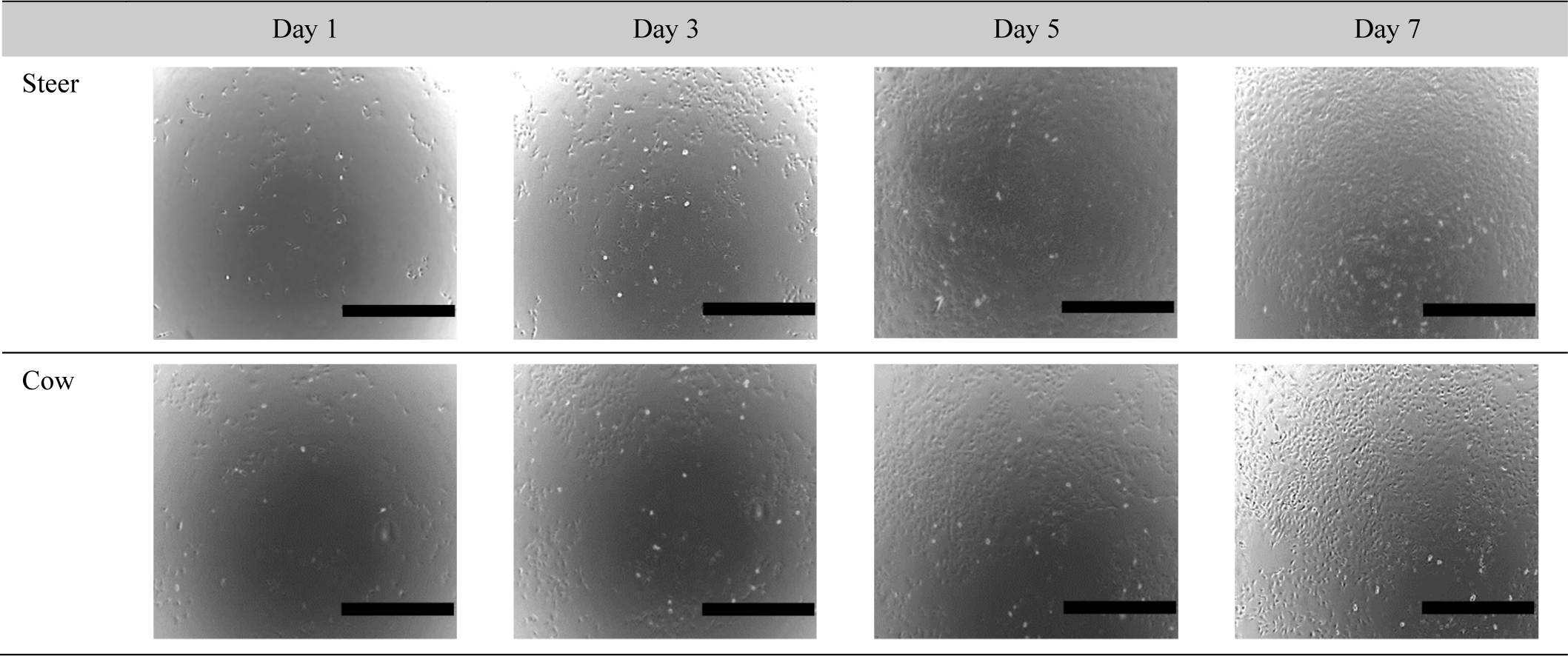
Each muscle contains SMSC, which play a key role in muscle development, proliferation, metabolic characteristics, and meat quality (Bazgir et al., 2016; Koike et al., 2022). SMSC, as the primary proliferative components of skeletal muscles, exhibit self-renewal capabilities that are crucial for muscle growth and repair. Moreover, the characteristics of muscle cells can vary depending on factors such as sex, breed, and age (Kim et al., 2022). These variations may further influence the overall metabolic characteristics and meat quality of the muscle. Approximately 35%–60% of the total muscle mass in livestock and fish is utilized for human food production. Among these, skeletal muscles comprise approximately 90% muscle fibers and 10% connective and adipose tissues (Listrat et al., 2016). The growth performance and body composition of cattle are influenced by sex. Comparative studies on growth performance based on sex have revealed that bulls exhibit higher growth rates than steers and heifers (Reyneke, 1976). In addition, studies in mice have indicated a greater number of myofiber SMSC in males than in females (Day et al., 2010). These findings confirmed the increased presence of myofiber SMSC based on hormone levels associated with sex. To this end, we examined whether there were sex-based differences in the growth rates of bovine muscle cells (Fig. 1). The results confirmed that steers exhibited higher cell growth rates than cows (Figs. 2A and B). In the case of steers, cell counts started at 2×104 and reached 6.93±0.251×104, 9.33±0.289×104, 10.83±0.285×104, and 13.5±0.5×104 during 7 days, while cows started at 2×104 and reached 4.56±0.115×104, 8.66±0.289×104, 9±0.284×104, and 11.3±0.763×104, respectively (Fig. 2A). As indicated in Fig. 2B, when calculating the doubling time using cell counts, significant differences were observed between steers and cows at 24, 120, and 168 h (p<0.0001). To confirm this finding, examination of cell images revealed that steers exhibited a higher cell growth rate and lower doubling time than cows, and this difference in muscle cell growth rate was also significantly influenced by sex.
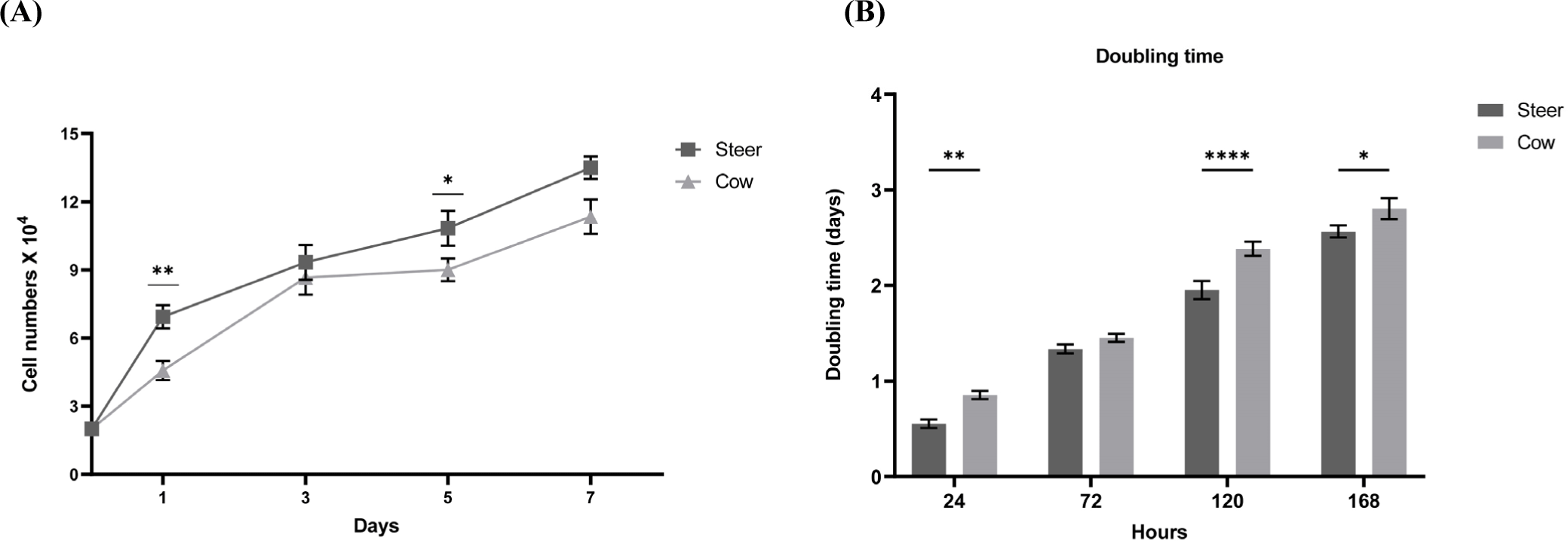
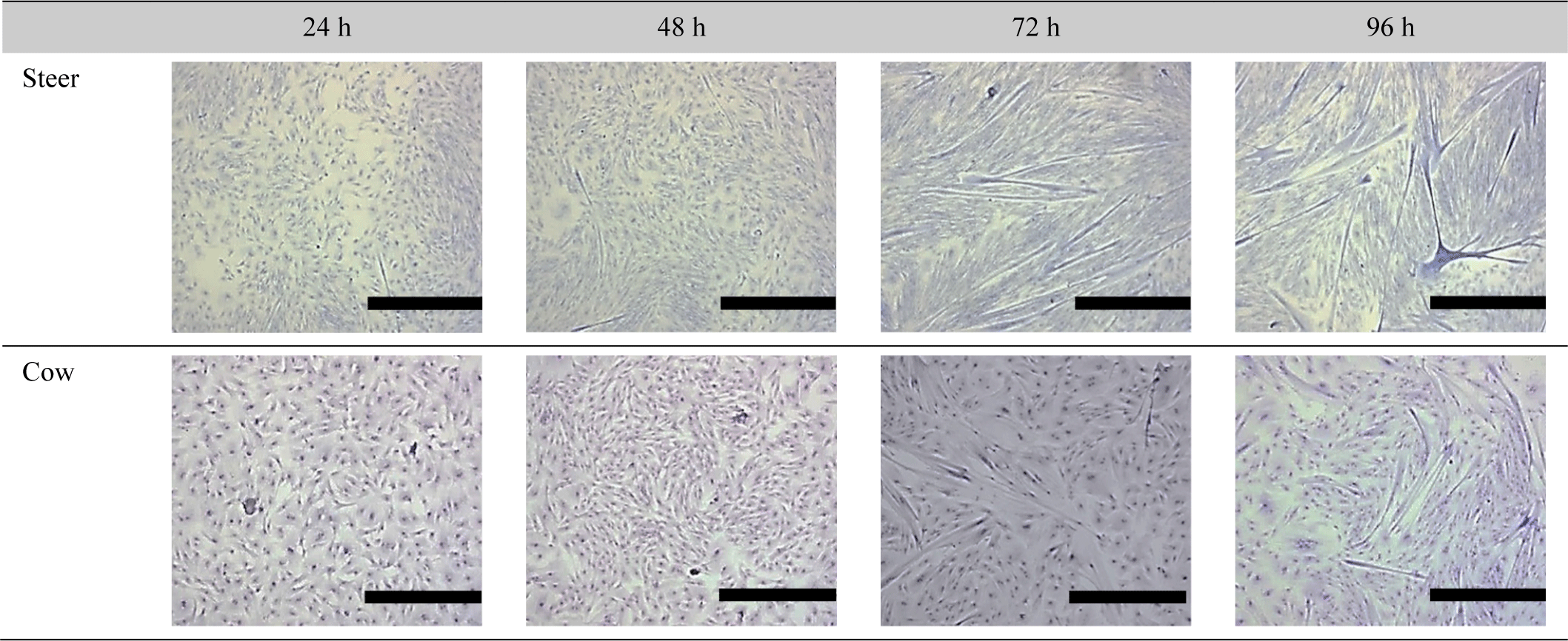
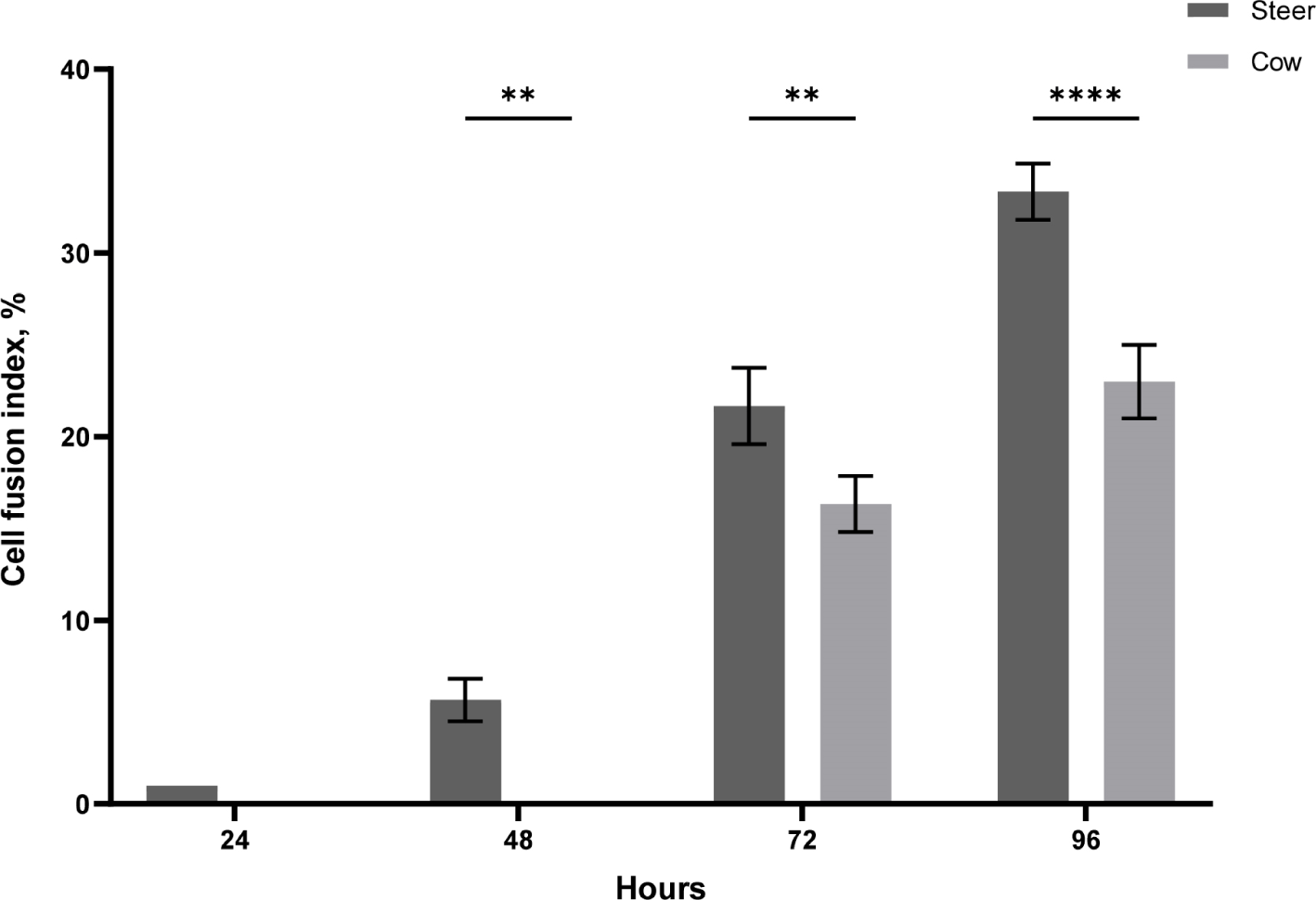
Verdijk et al. (2014) observed differences in muscle growth and differentiation rates between male and female muscle cells. Mouse experiments regarding sex-specific differentiation rates and growth indicated that male mice demonstrated faster differentiation and growth rates than female mice (Neal et al., 2012). This was confirmed to affect the differentiation rates based on doubling time. Moreover, Rosa-Caldwell and Greene (2019) stated that males tend to exhibit higher levels of testosterone, a transcription factor that enhances myogenesis and muscle protein synthesis. Additionally, the administration of testosterone to animals or individuals with low testosterone levels, such as elderly male rodents, results in increased muscle mass (Rosa-Caldwell and Greene, 2019; Sinha et al., 2014; Szeszycki, 1997). Based on previous studies, we investigated whether there are sex-based differences in the muscle satellite cell differentiation rate in cattle. For differentiation, the cell culture medium was replaced with DM when the cells were over 80% confluent, and the time point was set at 0 h. As shown in Fig. 3, the differentiation results between steers and cows revealed that steers had already formed myotubes after 24 h, whereas cows showed no differentiation. After 48 h, steers continued to undergo differentiation, while cows had not yet initiated the process. By 72 h, the differentiation of cow SMSC had begun, and steers formed a greater number of myotubes. After 96 h, both cow and steer muscle cells exhibited a gradual increase in differentiation. Fig. 4 displays the cell fusion index results for steer and cow SMSC. The steers exhibited fusion indices of 5.6% at 48 h, 21.66% at 72 h, and 33.3% at 96 h (p<0.001). In contrast, cows showed a fusion index of 0% at 48 h, 16.33% at 72 h, and 23% at 96 h (p<0.001). This differentiation pattern also exhibited similarities with proliferation, indicating sex-based differences in differentiation (Verdijk et al., 2014).
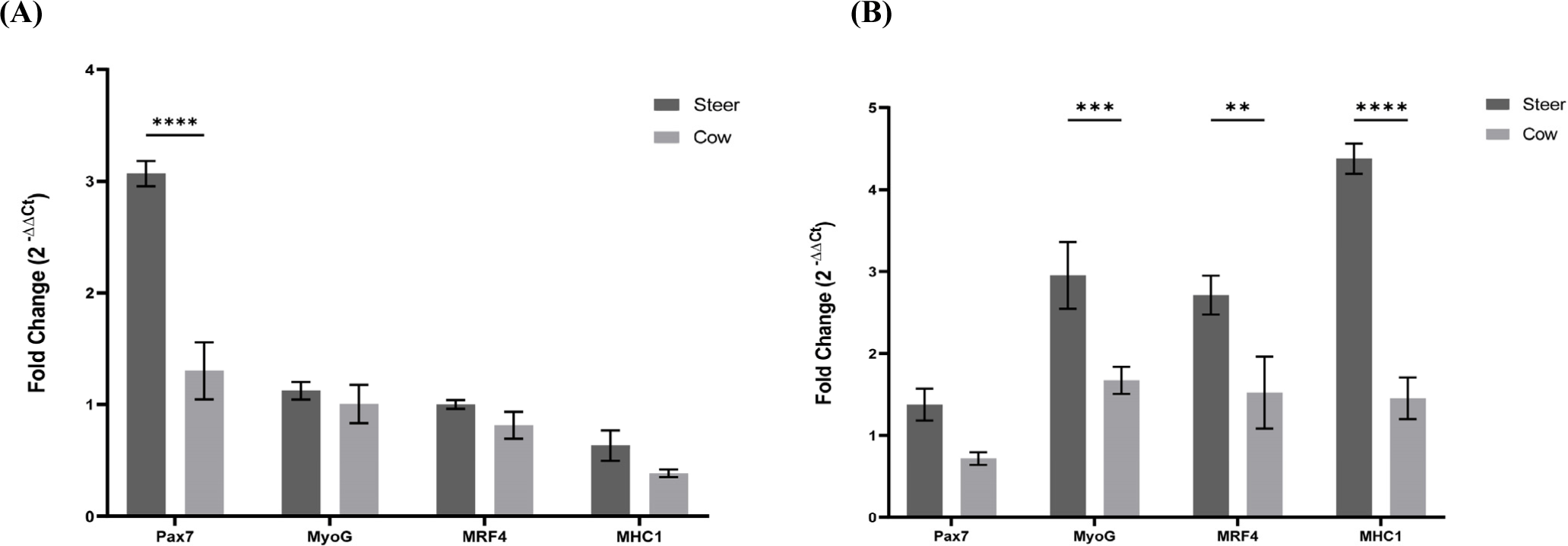
Skeletal muscles possess a high regenerative capacity, allowing them to adapt to physiological demands such as growth or exercise (Murach et al., 2021). In the quiescent state, SMSC are activated, expanded, and undergo differentiation following a program similar to fetal muscle formation. Muscle growth and differentiation occur in four stages: progenitor cells, myoblasts, myotubes, and myofibers (Choi et al., 2021). In the early progenitor stage, the Pax7 gene is expressed, playing a critical role in muscle growth and regeneration (Cosgrove et al., 2009). Activated SMSC are also involved in the repair of damaged muscles and the generation of new muscle fibers. Subsequently, in the myoblast stage, muscles grow until they differentiate. At this stage, Pax7 expression decreases and the myogenin gene begins to be expressed. Myogenin is crucial for muscle cell maturation, myotube formation, the promotion of muscle cell differentiation, and protein production (Zammit, 2017). During the transition from myocyte to myotube, MRF4 expression begins (Zammit, 2017). MRF4, another muscle differentiation gene, contributes to muscle cell maturation and myofiber formation (Kassar-Duchossoy et al., 2004). Activated SMSC play a pivotal role in regulating cell proliferation and differentiation to maintain muscle formation and function. Subsequently, they evolve into myofibers in which MHC1 is highly expressed. MHC1 is a protein associated with muscle contraction that enables muscle fibers to contract (Robelin et al., 1993). It is a key muscle protein that enables contraction within muscle cells. These genes cooperate during muscle development and maturation in order to regulate and maintain muscle formation and function. To understand the roles of these genes, we conducted real-time PCR analysis of the growth and differentiation of steers and cows. On the 5th day of growth in the gene expression comparison analysis (Fig. 5A), we observed that Pax7 was expressed at higher levels in steers than in cows (p<0.0001). MyoG, MRF4, and MHC1 showed a tendency for higher expression levels in steers than in cows; however, these differences were not statistically significant. In the gene expression comparison analysis on the 3rd day of differentiation (Fig. 5B), Pax7 showed lower expression levels than those during cell growth. MyoG, MRF4, and MHC1 exhibited higher expression levels in steers than in cows (p<0.001, p<0.01, and p<0.0001, respectively). These results support those of the experiments related to cell growth and differentiation, indicating that steers with high Pax7 expression have a higher cell count and lower doubling time for cell growth than cows. Additionally, during differentiation, steers with a higher expression of MyoG, MRF4, and MHC1 showed a higher fusion index than cows.
Conclusion
In summary, our study revealed significant differences in cellular proliferation and differentiation processes within the muscle tissue of Hanwoo steers and cows. Steers exhibited higher growth and fusion rates, reflected in the increased Pax7 expression during growth and elevated MyoG, MRF4, and MHC1 expression during differentiation, compared to cows (p<0.0001). These findings mirror observations in human muscle cells, indicating accelerated growth and differentiation, which are crucial for meat production. These investigations contribute to a better understanding of muscle tissue development and offer insights into the modulation of meat quality in the meat production industry.













