Introduction
Milk fatty acids act a part significant role in nutrition, contributing to a well-rounded and balanced diet. They provide a valuable source of energy and serve as carriers for fat-soluble vitamins, essential fatty acids, and another significant nutrients (Ahsani et al., 2022; Bayraktar and Shoshin, 2021; Samková et al., 2021). Milk fatty acids in dairy cattle can be categorized into several groups stand on their chemical structure and characteristics. Saturated fatty acids (SFAs) are fatty acids without any double bonds in their carbon chain and are mostly solid at room temperature. Monounsaturated fatty acids (MUFAs) have one double bond in their carbon chain and are typically liquid at room temperature (Dujková et al., 2015; Houaga et al., 2018; Shingfield et al., 2013). Polyunsaturated fatty acids (PUFAs) have two or more double bonds in their carbon chain and are essential fatty acids, meaning they cannot be synthesized by the body and must be obtained from the diet. Trans fatty acids are unsaturated fatty acids that have a specific configuration of double bonds and can occur naturally in small amounts in ruminant-derived foods like milk, but artificial trans fats created through industrial processes have been associated with negative health effects (Deckelbaum and Torrejon, 2012; Jenkins and McGuire, 2006; Shahidi and Ambigaipalan, 2018; Xiong et al., 2022). The genetic structure of cattle, including gene polymorphisms, can influence the fatty acid composition of their milk. Polymorphisms in genes related to lipid metabolism, such as stearoyl-coenzyme A desaturase (SCD) and fatty acid synthase (FASN), can impact the profile of fatty acids in milk. These gene polymorphisms can result in variations in the levels of specific fatty acids, altering the nutritional and functional properties of the milk (Bayraktar, 2022; Bayraktar and Özcan, 2023; Macciotta et al., 2008; Mele et al., 2007; Rincon et al., 2012).
The SCD gene encodes an enzyme that plays a critical role in fatty acid synthesis, converting SFAs to MUFAs. In cattle, this gene is vital for the regulation of fat composition in meat and milk, impacting livestock quality and yield. The primary function of SCD is the synthesis of cis-9, trans-11 conjugated linoleic acid and oleic acid (Alim et al., 2012; Bayraktar and Özcan, 2023; Cecchinato et al., 2012; Inostroza et al., 2013; Li et al., 2020). These fatty acids have several implications for human health and provide to the nutritional quality and stability of meat and dairy products. Variations in the SCD gene impact the fatty acid composition in cattle, affecting the quality of meat and milk produced. Bovine SCD gene is found on BTA26 chromosome. The bovine SCD gene is approximately 8.9 kb in length, consisting of six exons and five introns (Conte et al., 2010; Kgwatalala et al., 2009; Milanesi et al., 2008; Soltani-Ghombavani et al., 2016; Taniguchi et al., 2004). The FASN gene plays an essential role the synthesis of fatty acids in cattle. It encodes an enzyme that is a multi-domain protein involved in fatty acid biosynthesis, specifically in the conversion of acetyl-CoA and malonyl-CoA into palmitic acid (C16:0), a long-chain fatty acid (Morris et al., 2007; Roy et al., 2005; Zhang et al., 2008). This fatty acid is further elongated and desaturated to produce various kinds of fatty acids required for the growth, maintenance, and reproduction of cattle. The primary function of the FASN gene in cattle is to regulate the synthesis of fatty acids, which impacts milk production, fat deposition, and fatty acid profiles (Kumar et al., 2017; Mauric et al., 2019; Miluchová et al., 2023). FASN also influences meat and carcass fatness quality traits, essential factors for beef cattle breeding programs. Variations in the FASN gene may affect the meat and milk quality, production efficiency, and overall livestock yield. The bovine FASN gene is located on BTA19 chromosome. The bovine FASN gene spans approximately 44-45 kb in length, consisting of 42 exons and 41 introns (Li et al., 2012; Matsuhashi et al., 2011; Roy et al., 2001; Roy et al., 2006).
The objective of this study is to assess the impact of SCD and FASN gene polymorphisms on milk production and the composition of fatty acids in Holstein cows.
Materials and Methods
A sum of 100 cows were used from the Research and Applications Farm of Cukurova University Adana, Turkey. The cows in this study were provided with a total mix ration (TMR) as their feed, which had a concentrated feed-to-forage ratio of 60:40. The TMR formulation comprised of various ingredients, including corn silage, alfalfa, wheat straw, and a concentrate with a crude protein content of 19% and a metabolizable energy value of 2,700 kcal/kg.
Cows in their second and third lactation to collect raw milk samples. The specimens were specifically assembled during the forenoon milking from cows at a specific stage of lactation, between 70 and 80 days in milk. To preserve the milk for subsequent fatty acid analysis, a portion of each raw milk sample (150 mL) was frozen at a temperature of –80°C and stored until further analysis.
The lipid analysis was conducted following a modified version of the method developed by Bligh and Dyer (1959). A total of 15 g of the sample was mixed with 120 mL of a methanol/chloroform solution (1:2 v/v) using a T25 Ultraturrax (Ika-Werke, Staufen, Germany) for 2 min. Next, 20 mL of a 0.4% CaCl2 solution was added to the mixture. The homogenate was filtered through a Whatman filter paper (Schleicher & Schuell, Dassel, Germany; 5951/2 185 mm), and the filtrate was transferred to a separatory funnel for phase separation. The chloroform-lipid fraction was collected and transferred to a rotary evaporator for evaporation under vacuum. The flasks containing the lipid extracts were then placed in an oven at 90°C for 1 hour, followed by cooling in a desiccator. Finally, the vials containing the lipid extracts were weighed using a precision balance with an accuracy of 0.1 mg.
The fatty acid methyl esters (FAMEs) of the extracted lipid were prepared following the method described by Ichihara et al. (1996). For this, 25 mg of the extracted oil sample was combined with 4 mL of 2M KOH and 2 mL of n-heptane. The mixture was then vortexed for 2 min at room temperature to ensure thorough mixing. Subsequently, the mixture was centrifuged at 1,930×g for 10 min, and the upper heptane layer was carefully picked up for further analysis using gas chromatography (GC).
The configuration of fatty acids was analyzed using a GC Clarus 500 device from Perkin-Elmer (Waltham, MA, USA). The analysis employed a flame ionization detector and an SGE column (60 m×0.32 mm ID BP×70×0.25 μm) from either the USA or Australia. The identification of fatty acids was accomplished by comparing the retention times of the FAMEs from Supelco (Bellefonte, PA, USA; Catalogue No: 18919) with a standard 37-component FAME mixture. To ensure accuracy, three replicates of the GC analysis were performed, and the results were expressed as the mean±SE in terms of GC area %.
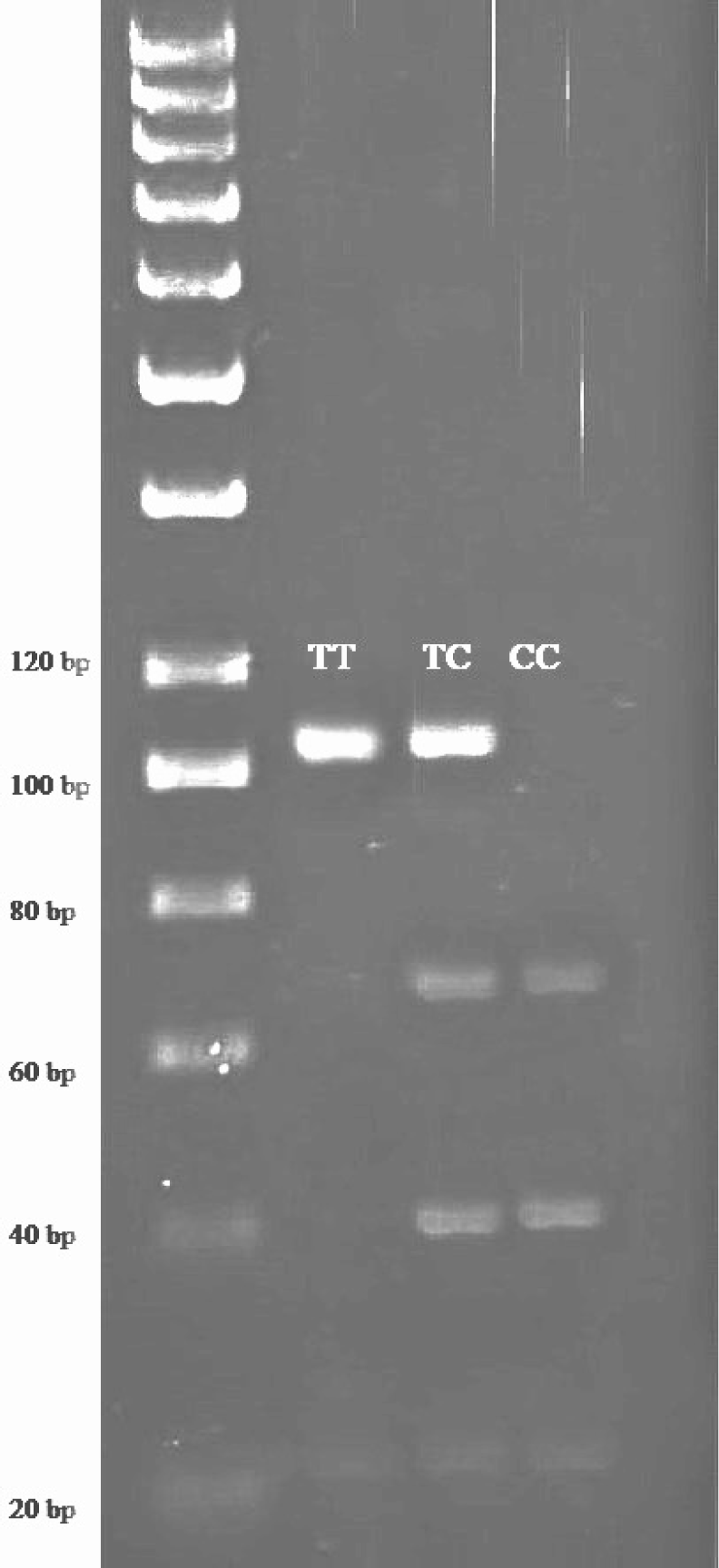
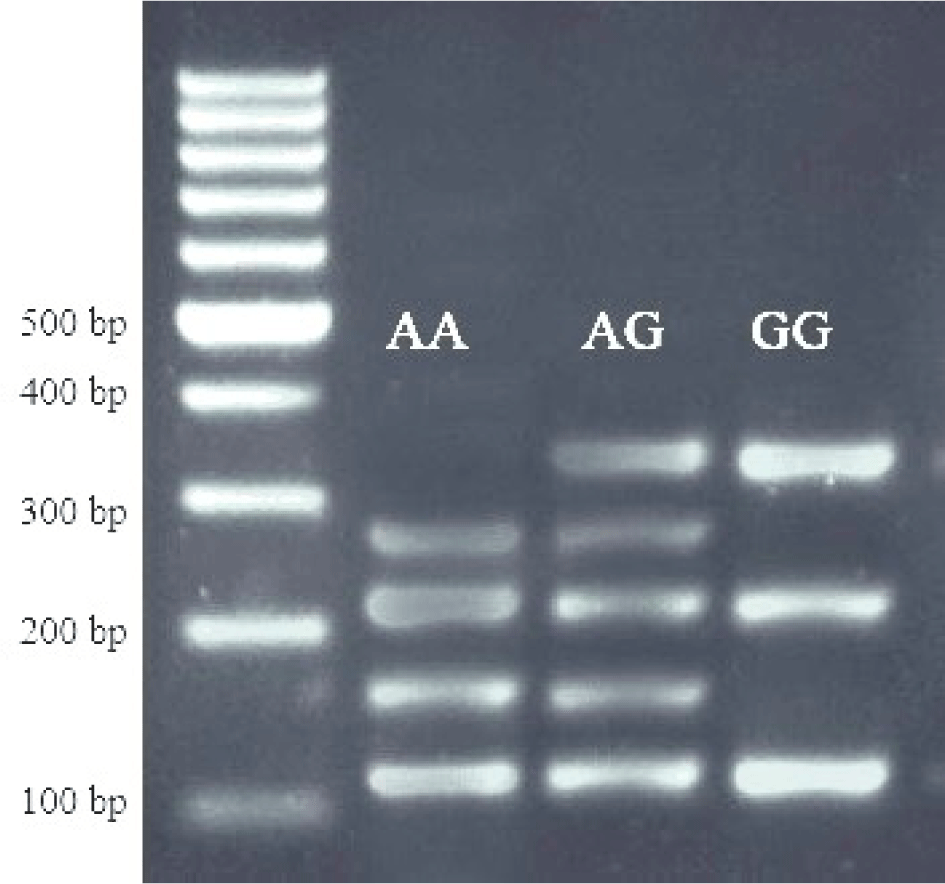
Blood specimens were obtained from the vena jugularis for DNA analysis. Genomic DNA was extracted from the whole blood using a modified salting-out process based on the method identified by Miller et al. (1988). The primer sequences used in this study were designed by Inostroza et al. (2013). The primer sequences for the SCD gene locus were 5'-ACCTG GCTGGTGAATAGTGCT-3' and 5'-TCTGGCACGTAACCTAATACCCT-3'. The PCR product length was 170 bp. The restriction enzyme used was SatI. After digestion, the genotype patterns were obtained as 122, 28, and 20 bp (TT); 122, 75, 47, 28, and 20 bp (TC); and 75, 47, 28, and 20 bp (CC). The PCR conditions included an initial denaturation at 95°C for 5 min, followed by 30 cycles of denaturation at 95°C for 20 s, annealing at 61°C for 20 s, and extension at 72°C for 30 s. A final extension was performed at 72°C for 10 min. The mutation was located at the 5th exon, with a nucleotide change of 878T>C. The primer sequences for the FASN gene locus were 5'-TGCTGTCCATGCGGGAAATC-3' and 5'-TGAGCACAT CTCGAAAGCCA-3'. The PCR product length was 758 bp. The restriction enzyme used was MscI. After digestion, the genotype patterns were obtained as 273, 217, 167, and 101 bp (AA); 440, 273, 217, 167, and 101 bp (AG); and 440, 217, and 101 bp (GG). The PCR conditions included an initial denaturation at 95°C for 5 min, followed by 30 cycles of denaturation at 95°C for 20 s, annealing at 58°C for 20 s, and extension at 72°C for 30 s. A final extension was performed at 72°C for 10 min. The mutation was located at the 38th exon, with a nucleotide change of g.17924A>G. A reaction volume of 20 μL was prepared for PCR amplification. Included were 5 μL of DNA template, 5 μL of 2X Dream Taq Green PCR Master Mix, 0.30 μL of each primer, and 9.4 μL of distilled water.
The PCR products were subjected to digestion using a fast-digest enzyme (Thermo Fisher Scientific, Waltham, MA, USA). The digestion reaction mixture consisted of 5 μL of PCR product, 8.5 μL of distilled water, 1 μL of 10X buffer, and 0.5 μL of the restriction enzyme, resulting in a total volume of 15 μL. The digestion products were then separated on a 3% agarose gel using an electrophoresis system. The gel was run at 85 V for 50 min in 0.5X TBE buffer. To visualize the DNA bands, the gel was stained with ethidium bromide. The gel was examined under ultraviolet lights to observe and analyze the resulting DNA fragments.
To analyze the genetic data, the software program POPGENE (version 1.32) was utilized. This software was employed to calculate genotype frequencies, allele frequencies, and conduct a χ2 test to assess the Hardy-Weinberg equilibrium at the gene locus. For the statistical analysis of the association between individual genotypes and milk production, as well as milk fatty acid components, Minitab 19 software was employed. The data obtained from the analysis were presented as “mean±SE” to provide a comprehensive representation of the results. To determine the association, the general linear model was applied using a specific statistical model as outlined below. This model was chosen to assess the relationship between genotypes and the studied variables. Tukey multiple range test was tested for mean differences among subclasses.
Yijk: phenotypic value; μ: overall means; αi: effect of genotypes; βj: effect of parity (two and three); γk: year and season of the first calving; Єij: random errors.
Results and Discussion
The genotype frequencies observed for the SCD gene were as follows: TT (0.21), TC (0.44), and CC (0.35; Table 1 and Fig. 1). The corresponding allele frequencies were T (0.43) and C (0.57). The χ2 test for hardy-weinberg equilibrium showed a value of 1.048 with a p-value of 0.547, indicating that the genotype distribution of the SCD locus is in agreement with hardy-weinberg equilibrium expectations.
| Gene locus | Genotype frequency | Allele frequency | χ2 | p-value |
|---|---|---|---|---|
| SCD | TT (0.21) TC (0.44) CC (0.35) |
T (0.43) C (0.57) |
1.048 | 0.547 |
| FASN | AA (0.15) AG (0.35) GG (0.50) |
A (0.33) G (0.67) |
4.092 | 0.384 |
For the FASN gene, the genotype frequencies were AA (0.15), AG (0.35), and GG (0.50). The allele frequencies were A (0.33) and G (0.67; Fig. 2). The χ2 test for hardy-weinberg equilibrium showed a value of 4.092 with a p-value of 0.384, suggesting that the FASN locus also conformed to hardy-weinberg equilibrium.
About SFA, the fatty acid composition of C4:0, and C11:0 showed significant associations with the SCD gene polymorphism. In the case of C4:0, the highest average was registered in the TT genotype (3.2701±0.023; p=0.023; Table 2). Similarly, for C11:0, the highest average was observed in the TC genotype (0.35267±0.0065; p=0.047). For MUFA, specifically C18:1 trans6, the TT genotype exhibited the highest average (0.37547±0.006; p=0.043). For PUFA, C18:3n3 displayed a significant association with the TT genotype showing the highest average (0.4738±0.015; p=0.029). For C20:2, the highest average was also found in the TT genotype (0.14546±0.0058; p=0.044).
The fatty acid composition is significantly associated with the FASN gene polymorphism in the context of certain fatty acid types (p<0.05; Table 3). For the SFA, the C14:0 showed significant associations with the FASN gene polymorphism, specifically having the highest average in the genotype AA (14.034±0.15; p=0.022). In the context of MUFA, C18:1 trans9, C18:1 trans11, and C24:1 n9 showed significant associations with the FASN gene polymorphism. For C18:1 trans9, the genotype AA had the highest average (0.22849±0.0079; p=0.028). For C18:1 trans11, the highest average was found in genotype GG (1.4737±0.012; p=0.030). As for C24:1 n9 the highest average was reported for the genotype AG (1.2564± 0.036; p=0.002). As for the PUFA, C18:2n6, C18:3n3, and C22:6 n3 showed significant associations with the FASN gene polymorphism. The fatty acid C18:2n6 showed the highest average in genotype AA (0.4475±0.027; p=0.034). For C18:3n3, the highest average was found in genotype AA (0.4653±0.017; p=0.013), and for C22:6 n3, the highest average was seen in genotype AA (0.21220±0.0093; p=0.004).
The present study analyzed the genotype distribution of the SCD and FASN gene loci and their association with milk fatty acid composition in Holstein cows. The genotype distribution of both gene loci was found to be in Hardy-Weinberg Equilibrium, indicating no deviations from expected frequencies based on random mating. These findings provide a solid foundation for further analysis of the genetic variations and their relationships with milk fatty acid profiles in Holstein cattle. Regarding the SCD gene locus, our results revealed two different alleles C and T with frequency 0.57 and 0.43 respectively. The genotypes frequency were 0.35 (CC), 0.44 (TC) and 0.21 (TT). These findings are consistent with previous studies by Bayraktar and Özcan (2023) and Safina et al. (2020), which also reported similar genotype distributions in Holstein cattle. Furthermore, our study demonstrated significant relationships between the SCD gene polymorphism and specific fatty acid profiles, including C4:0, C11:0, C18:1 trans6, C18:3n3, and C20:2. The TT and TC genotypes exhibited the highest averages for these fatty acids. These results align with the observations made by Inostroza et al. (2013) and Li et al. (2020), who reported associations between SCD gene polymorphism and altered fatty acid composition in milk. In the case of the FASN gene locus, our study identified three genotypes: AA, AG, and GG, with frequencies of 0.15, 0.35, and 0.50, respectively. The allele frequencies for A and G were 0.33 and 0.67, respectively. These findings are comparable to the studies conducted by Mauric et al. (2019) and Miluchová et al. (2023), which reported similar genotype distributions in different cattle populations. We found significant associations between FASN gene polymorphism and several fatty acid profiles, including SFAs, MUFAs, and PUFAs. Notably, fatty acids such as C14:0, C18:1 trans9, C18 trans11, C24:1 n9, C18:2n6, C18:3n3, and C22:6 n3 exhibited prominent prevalence in the AA, GG, and AG genotypes. These findings align with the observations made by Inostroza et al. (2013) and Kumar et al. (2017), who reported associations between FASN gene polymorphism and altered milk fatty acid composition. Comparisons with previous studies strengthen the significance of our findings and provide a broader context for understanding the genetic influences on milk fatty acid composition. Several studies, including Bayraktar and Özcan (2023), Macciotta et al. (2008), and Mele et al. (2007), have reported associations between SCD gene polymorphism and milk production traits. Our study further contributes to this knowledge by demonstrating associations between specific SCD genotypes and altered fatty acid profiles. Similarly, previous studies such as Houaga et al. (2018), Rincon et al. (2012), and Roy et al. (2006) have highlighted the influence of SCD gene polymorphism on milk fat composition, supporting our findings. In the case of the FASN gene, studies by Inostroza et al. (2013) and Mauric et al. (2019) have reported associations between FASN gene polymorphism and altered milk fatty acid content. Our findings align with these studies, further emphasizing the role of FASN gene polymorphism in shaping milk fatty acid profiles. Moreover, Miluchová et al. (2023) identified an association between FASN gene polymorphism and milk produce and composition, which complements our observations. Our study provides valuable insights into the associations between SCD and FASN gene polymorphisms and milk fatty acid composition in Holstein cows. The findings support previous studies and reinforce the potential role of these gene polymorphisms as markers for modifying fatty acid composition. Understanding the genetic influences on milk fatty acid profiles can inform breeding strategies to improve milk quality and develop dairy products with specific nutritional attributes. Further research is warranted to explore additional genetic markers and their interactions in order to enhance our understanding of the genetic regulation of milk fatty acid composition.
Conclusion
In this practice, we investigated the allele and genotype frequencies of the SCD and FASN genes and their associations with fatty acid composition. These findings suggest that the genetic variations in the SCD and FASN genes are associated with milk fatty acid compositions in our study population. Understanding these associations can provide insights into the genetic mechanisms underlying fatty acid metabolism and may have implications for nutritional and health-related aspects. Further research is warranted to validate these findings in larger and more diverse populations and to explore the functional implications of these genetic associations.













