SHORT COMMUNICATION
Monitoring mRNA Expression Patterns in Macrophages in Response to Two Different Strains of Probiotics
Sang-Pil Choi1,†
,
Si-Won Park1,†
,
Seok-Jin Kang1
,
Seul Ki Lim2
,
Min-Sung Kwon2
,
Hak-Jong Choi2,*
,
Taehoon Chun1,*
Author Information & Copyright ▼
1Department of Biotechnology, College of Life Sciences and Biotechnology, Korea University, Seoul 02841, Korea
2Technology Innovation Research Division, World Institute of Kimchi, Gwangju 61755, Korea
*Corresponding author: Hak-Jong Choi, Technology Innovation Research Division, World Institute of Kimchi, Gwangju 61755, Korea, Tel: +82-62-610-1729, Fax: +82-62-610-1850, E-mail:
hjchoi@wikim.re.kr
*Corresponding author: Taehoon Chun, Department of Biotechnology, College of, Life Sciences and Biotechnology, Korea University, Seoul 02841, Korea, Tel: +82-2-3290-3069, Fax: +82-2-3290-3507, E-mail:
tchun@korea.ac.kr
† These authors contributed equally to this work.
© Korean Society for Food Science of Animal Resources. This is an Open-Access article distributed under the terms of the
Creative Commons Attribution Non-Commercial License (http://creativecommons.org/licenses/by-nc/3.0) which permits
unrestricted non-commercial use, distribution, and reproduction in any
medium, provided the original work is properly cited.
Received: Apr 27, 2023 ; Revised: Jun 01, 2023 ; Accepted: Jun 05, 2023
Published Online: Jul 01, 2023
Abstract
As an initial study to elucidate the molecular mechanism of how probiotics modulate macrophage activity, we monitored mRNA expression patterns in peritoneal macrophages (PMs) treated with two different strains of probiotics. After treatment with either Weissella cibaria WIKIM28 or Latilactobacillus sakei WIKIM50, total RNAs from PMs were isolated and subjected into gene chip analyses. As controls, mRNAs from vehicle (phosphate-buffered saline, PBS)-treated PMs were also subjected to gene chip analysis. Compared to vehicle (PBS)-treated PMs, WIKIM28-treated and WIKIM50-treated PMs exhibited a total of 889 and 432 differentially expressed genes with expression differences of at least 4 folds, respectively. Compared to WIKIM28-treated PMs, WIKIM50-treated PMs showed 25 up-regulated genes and 21 down-regulated genes with expression differences of more than 2 folds. Interestingly, mRNA transcripts of M2 macrophage polarization marker such as anxa1, mafb, and sepp1 were increased in WIKIM50-treated PMs comparing to those in WIKIM28-treated PMs. Reversely, mRNA transcripts of M1 macrophage polarization marker such as hdac9, ptgs2, and socs3 were decreased in WIKIM50-treated PMs comparing to those in WIKIM28-treated PMs. In agreement with these observations, mRNA expression levels of tumor necrosis factor-α and interleukin-1α were significantly reduced in WIKIM50-treated macrophages compared to those in WIKIM28-treated macrophages. These results may indicate that probiotics can be classified as two different types depending on their ability to convert macrophages into M1 or M2 polarization.
Keywords: differentially expressed gene; macrophage; probiotics
Introduction
Colonization of selective probiotics in gastrointestinal tract might lead to immune modulation within gut microenvironment by communications with mucosal immune cells (Belkaid and Hand, 2014). Furthermore, the crosstalk between probiotics and immune cells could affect the status of various physiological conditions of humans (Kumari et al., 2021). According to this, several connections between gut and specific organs such as gut-brain, gut-liver, and gut-lung axes have been proposed to explain the functional importance of selective probiotics (Dumas et al., 2018; Powell et al., 2017; Tripathi et al., 2018).
Probiotics can modulate immune responses by mainly interacting with pattern recognition receptors (PRRs) expressed on professional antigen presenting cells (pAPCs) or epithelial cells (Fukata and Arditi, 2013). After interacting with PRRs, specific receptor-mediated signal transduction pathway can turn on and change the phenotype of pAPCs (Fukata and Arditi, 2013). Besides dendritic cells, macrophages are a major population of pAPCs that can modulate inflammatory responses by either triggering inflammation or suppressing inflammation for tissue repair (Lawrence and Natoli, 2011). Therefore, two types of macrophages, inflammatory macrophages (M1 macrophages) and anti-inflammatory macrophages (M2 macrophages), have been proposed depending on their functions (Lawrence and Natoli, 2011). Recently, many observations have demonstrated that M1 or M2 macrophage skewing conditions might affect several pathologic conditions in human diseases (Shapouri-Moghaddam et al., 2018).
Among probiotics, Weissella cibaria is considered as a good starter of Kimchi fermentation because it has many beneficial effects including anti-microbial activities, antioxidant activities and immunomodulatory effects (Lim et al., 2017; Yu et al., 2018; Zhu et al., 2022). Latilactobacillus sakei is also used for the food fermentation or preservation. Especially, L. sakei is known for a commercial meat starter of meat fermentation (Leroy et al., 2006). Therefore, W. cibaria and L. sakei are good resources for food industry.
In the present study, we profiled mRNA expression patterns in macrophages after treatment with two different strains of probiotics isolated from Kimchi, W. cibaria WIKIM28 and L. sakei WIKIM50 (Lim et al., 2017). Results of this study might provide valuable data to understand the molecular mechanisms of macrophage polarization triggered by different types of probiotics.
Materials and Methods
Probiotics
Isolation and species identification of W. cibaria WIKIM28 from Gat Kimchi have been described previously (Lim et al., 2017). WIKIM50 was isolated from Baechu Kimchi and identified as L. sakei based on 16S rRNA gene sequencing according to a previously established method (Lim et al., 2017). WIKIM28 and WIKIM50 were sub-cultured more than 10 times before treating peritoneal macrophages (PMs) for RNA-Seq data analyses.
Stimulation of peritoneal macrophages (PMs) by probiotics
PMs were isolated according to a previously established method (Pineda-Torra et al., 2015). Briefly, 3% thioglycolate (Becton Dickinson, Sparks, MD, USA) in phosphate-buffered saline (PBS) was intraperitoneally injected into C57BL/6 mice (Orient Bio, Seongnam, Korea) at 1 mL per mouse. On day 3, thioglycolate-injected mice were sacrificed to harvest peritoneal lavages. Cell pellets were then acquired after centrifuging peritoneal lavages at 400×g for 5 min at 4°C and resuspended in DMEM/F-12 (Thermo Fisher Scientific, Waltham, MA, USA) medium supplemented with 10% (v/v) fetal bovine serum and 1% penicillin/streptomycin (DMEM/F-12-10). After resuspension, cells were plated into tissue culture plates and incubated in a humidified 5% CO2 atmosphere at 37°C. After overnight culture, adherent cells were harvested and counted. The purity of PMs was validated by flow cytometry analyses using anti-mouse CD11b (eBioscience, San Diego, CA, USA) and anti-mouse F4/80 (BioLegend, San Diego, CA, USA) antibodies. More than 90% of cells were CD11b+F4/80+ cells (data not shown). After isolation, PMs (2×106 cell/well) were seeded into a 12-well plate in 1 mL of DMEM/F-12-10. Heat killed WIKIM28 or WIKIM50 (2×107 CFU/mL) was co-cultured with PMs in a humidified 5% CO2 atmosphere at 37°C for 24 hours. As controls, vehicle (PBS) was used to treat PMs without incubation with probiotics.
Gene chip analyses
Total RNAs were isolated from vehicle (PBS)-, WIKIM28-, or WIKIM50-treated PMs using TRIzol (Thermo Fisher Scientific) according to the manufacturer’s protocol. The quality and quantity of each RNA sample were measured using an ND-1000 spectrophotometer (NanoDrop, Wilmington, DE, USA) and an Agilent 2100 Bioanalyzer (Agilent Technologies, Santa Clara, CA, USA). Gene chip analyses were then performed using Agilent SurePrint G3 Mouse gene expression V2 microarrays (8×60K) according to the manufacturer’s protocol (Agilent Technology, V 6.5, 2010). Briefly, total RNAs from each sample were linearly amplified and labeled with Cy3-dCTP. These labeled cRNAs were then purified with an RNAeasy mini kit (Qiagen, Hilden, Germany). The concentration and specific activity of the labeled cRNAs (pmol Cy3/μg cRNA) were measured with an ND-1000 spectrophotometer. For hybridization of microarray, each labeled cRNA was fragmented by adding 5 μL of 10×blocking agent and 1 μL of 25×fragmentation buffer and then heated at 60°C for 30 min. After heating, 25 μL of 2×hybridization buffer was added to dilute the labeled cRNA. Subsequently, 40 μL of hybridization solution was loaded into the gasket slide and assembled to microarray slides (Agilent Technologies). After assembling, microarray slides were incubated at 65°C in an Agilent hybridization oven (Agilent Technologies) for 17 hours. After incubation, microarray slides were washed at room temperature using washing buffer provided by manufacturer (Agilent Technologies). The hybridized array was then immediately scanned with an Agilent Microarray Scanner D (Agilent Technologies).
Raw data were extracted using a Feature Extraction Software (v11.0.1.1) provided by Agilent (Agilent Technologies). Comparative analysis between test sample and control sample was carried out using paired t-test. False discovery rate was controlled by adjusting p-value using Benjamini-Hochberg algorithm (Benjamini and Hochberg, 1995). For a differentially expressed genes (DEG) set, hierarchical cluster analysis was performed using complete linkage and Euclidean distance as a measure of similarity.
Transcriptional analysis by quantitative reverse-transcriptase real-time PCR (qRT-PCR)
To confirm gene chip analyses, qRT-PCR were conducted as described previously (Choi et al., 2018). Briefly, total RNAs from vehicle (PBS)-, WIKIM28-, or WIKIM50-treated PMs were isolated for synthesizing cDNAs using a First-Strand cDNA Synthesis Kit with oligo-dT primers (SuperScript RT; Thermo Fisher Scientific). Subsequently, quantitative real-time PCR was performed using one μg of each total RNA with QGreenTM 2X qPCR Master Mix (GenDEPOT, Katy, TX, USA) on a Bio-Rad CFX96 Real-Time Detection System (Bio-Rad, Hercules, CA, USA). Expression of each mRNA transcript was compared to the expression level of mouse Gapdh to obtain relative gene expression. Primer sequences for mouse gapdh were 5′-ATGGTGAAGGTCGGTGTGAA-3′ (sense) and 5′-GGTCGTTGATGGCAACAATCTC-3′ (anti-sense), resulting in a 100 bp product (Kang et al., 2022). PCR primers of M1 or M2 marker genes used in this study are listed in Table 1.
Table 1.
Primer sets of M1 or M2 macrophage signature genes analyzed by quantitative real-time reverse transcriptase-PCR (qRT-PCR)
| Gene name |
Classification |
Primer sequence (5′→ 3′) |
Tm (°C) |
Product size |
Accession number |
|
anxa1
|
M2 macrophage marker |
F: AGCTCTGGATCTGGAACTGA |
55.4 |
103 |
NM_010730.2 |
| R: TTCGTACAGCTTCTCGGCAA |
55.4 |
|
mafb
|
M2 macrophage marker |
F: GCGAGCAACTACCAGCAGAT |
57.4 |
125 |
NM_010658 |
| R: GCACTACGGAAGCCGTCGAA |
59.5 |
|
sepp1
|
M2 macrophage marker |
F: GAGCATCTTGGCAGCAGTAA |
55.4 |
140 |
NM_001042614 |
| R: GTGGTGTCTCAGCTCTCTAA |
55.4 |
|
hdac9
|
M1 macrophage marker |
F: ATCAGCTCAGTGGACGTGAA |
55.4 |
103 |
NM_024124 |
| R: GATCCACCACAGGCATCATC |
57.4 |
|
ptgs2
|
M1 macrophage marker |
F: ATCAGGTCATTGGTGGAGAGG |
58.2 |
137 |
NM_011198 |
| R: CATCAGACCAGGCACCAGA |
57.3 |
|
socs3
|
M1 macrophage marker |
F: CCTCAAGACCTTCAGCTCCAA |
58.2 |
82 |
NM_007707 |
| R: CCAGTAGAATCCGCTCTCCT |
57.4 |
Download Excel Table
Statistical analyses
Statistical significance of independent variable values was evaluated by Student’s t-test. Significant differences at a confidence level of 95%, 99%, 99.9% are labeled on each graph and indicated by asterisks ‘*’, ‘**’, and ‘***’, respectively.
Results and Discussion
Differentially expressed genes (DEGs) in peritoneal macrophages (PMs) treated with either WIKIM28 or WIKIM50
Compared with vehicle (PBS)-treated PMs, PMs treated with WIKIM28 and WIKIM50 showed a total of 889 and 432 DEGs with expression differences of at least 4 folds, respectively. Among them, 652 from WIKIM28-treated cells and 375 genes from WIKIM50-treated cells were up-regulated compared to those in vehicle (PBS)-treated PMs. Contrarily, 237 genes from WIKIM28-treated cells and 57 genes from WIKIM50-treated cells were down-regulated compared to those in vehicle (PBS)-treated cells.
DEGs with at least 2-fold expression difference in WIKIM50-treated cells compared to those in WIKIM28-treated cells are listed in Table 2. Among them, 25 genes were up-regulated, and 21 genes were down-regulated in WIKIM50-treated cells compared to those in WIKIM28-treated cells. Interestingly, several M2 macrophage marker genes such as anxa1, mafb, and sepp1 were up-regulated in WIKIM50-treated cells, whereas several M1 macrophage marker genes such as hdac9, ptgs2, and socs3 were down-regulated in WIKIM50-treated cells compared with those in WIKIM28-treated cells (Arnold et al., 2014; Barrett et al., 2015; Kim, 2017; Liu et al., 2021; Lu et al., 2017; Moraes et al., 2017). These results indicate that WIKIM28-treated macrophages are more likely to be polarized into M1 phenotype whereas WIKIM50-treated macrophages might exhibit M2 phenotype. Supporting this idea, mRNA transcripts of pro-inflammatory cytokines such as tumor necrosis factor-α and IL-1α were significantly decreased in WIKIM50-treated macrophages compared to those in WIKIM28-treated macrophages (Table 2).
Table 2.
DEGs between WIKIM28-treated macrophages and WIKIM50-treated macrophages1)
| Gene symbol |
Gene description |
Fold increase |
Accession number |
| Up-regulated genes in WIKIM50-treated macrophages compared to those in WIKIM28 |
|
cd55
|
CD55 antigen |
4.04 |
NM_010016 |
|
card11
|
Caspase recruitment domain family, member 11 |
3.97 |
NM_175362 |
|
rgs2
|
Regulator of G-protein signaling 2 |
3.64 |
NM_009061 |
|
cxcr3
|
Chemokine (C-X-C motif) receptor 3 |
3.35 |
NM_009910 |
|
mafb
|
v-maf musculoaponeurotic fibrosarcoma oncogene family, protein B (avian) |
2.99 |
NM_010658 |
|
tlr12
|
Toll-like receptor 12 |
2.79 |
NM_205823 |
|
nlrp1b
|
NLR family, pyrin domain containing 1B |
2.78 |
NM_001040696 |
|
mrc1
|
Mannose receptor, C type 1 |
2.74 |
NM_008625 |
|
nfatc2
|
Nuclear factor of activated T cells, cytoplasmic, calcineurin dependent 2 |
2.63 |
NM_001136073 |
|
cd93
|
CD93 antigen |
2.62 |
NM_010740 |
|
il6ra
|
Interleukin 6 receptor, alpha |
2.51 |
NM_010559 |
|
thbs1
|
Thrombospondin 1 |
2.47 |
NM_011580 |
|
st6gal1
|
Beta galactoside alpha 2,6 sialyltransferase 1 |
2.45 |
NM_145933 |
|
pparg
|
Peroxisome proliferator activated receptor gamma |
2.36 |
NM_011146 |
|
ccl6
|
Chemokine (C-C motif) ligand 6 |
2.34 |
NM_009139 |
|
c3ar1
|
Complement component 3a receptor 1 |
2.33 |
NM_009779 |
|
sepp1
|
Selenoprotein P, plasma, 1 |
2.22 |
NM_001042614 |
|
anxa1
|
Annexin A1 |
2.12 |
NM_010730 |
|
ccr5
|
Chemokine (C-C motif) receptor 5 |
2.12 |
NM_009917 |
|
masp1
|
Mannan-binding lectin serine peptidase 1 |
2.07 |
XM_006521828 |
|
iqgap3
|
IQ motif containing GTPase activating protein 3 |
2.04 |
NM_001033484 |
|
cd300lb
|
CD300 antigen like family member B |
2.02 |
NM_199221 |
|
alox5
|
Arachidonate 5-lipoxygenase |
2.01 |
NM_009662 |
|
celf2
|
CUGBP, Elav-like family member 2 |
2.01 |
NM_010160 |
|
clec9a
|
C-type lectin domain family 9, member a |
2.00 |
NM_001205363 |
| Down-regulated genes in WIKIM50-treated macrophages compared to those in WIKIM28 |
|
cxcl2
|
Chemokine (C-X-C motif) ligand 2 |
–22.80 |
NM_009140 |
|
cxcl1
|
Chemokine (C-X-C motif) ligand 1 |
–17.74 |
NM_008176 |
|
ptgs2
|
Prostaglandin-endoperoxide synthase 2 |
–11.32 |
NM_011198 |
|
cav1
|
Caveolin 1, caveolae protein |
–9.92 |
NM_001243064 |
|
cxcl5
|
Chemokine (C-X-C motif) ligand 5 |
–5.33 |
NM_009141 |
|
tnf
|
Tumor necrosis factor |
–5.25 |
NM_013693 |
|
il1a
|
Interleukin 1 alpha |
–4.74 |
NM_010554 |
|
hdac9
|
Histone deacetylase 9 |
–4.64 |
NM_024124 |
|
dnmt3l
|
DNA (cytosine-5-)-methyltransferase 3-like |
–4.51 |
NM_019448 |
|
nlrp1a
|
NLR family, pyrin domain containing 1A |
–3.35 |
NM_001004142 |
|
maff
|
v-maf musculoaponeurotic fibrosarcoma oncogene family, protein F (avian) |
–3.30 |
NM_010755 |
|
cxcl10
|
Chemokine (C-X-C motif) ligand 10 |
–3.23 |
NM_021274 |
|
il20rb
|
Interleukin 20 receptor beta |
–3.21 |
NM_001033543 |
|
ccl3
|
Chemokine (C-C motif) ligand 3 |
–3.10 |
NM_011337 |
|
cd40
|
CD40 antigen |
–3.00 |
NM_011611 |
|
ccl7
|
Chemokine (C-C motif) ligand 7 |
–2.96 |
NM_013654 |
|
il10
|
Interleukin 10 |
–2.94 |
NM_010548 |
|
socs1
|
Suppressor of cytokine signaling 1 |
–2.80 |
NM_009896 |
|
socs3
|
Suppressor of cytokine signaling 3 |
–2.72 |
NM_007707 |
|
rgs16
|
Regulator of G-protein signaling 16 |
–2.57 |
NM_011267 |
|
cd38
|
CD38 antigen |
–2.40 |
NM_007646 |
Download Excel Table
Besides genes involved in macrophage polarization, genes encoding several cell surface proteins including PRRs, chemokine receptors, cytokine receptors were also identified as DEGs (Table 2). However, there was no clear physiological relevance in these DEGs.
Validation of gene expression patterns by quantitative reverse-transcriptase real-time PCR (qRT-PCR)
To validate gene chip data, mRNA expression levels of DEGs related to macrophage polarization were monitored by qRT-PCR (Fig. 1). Results of qRT-PCR analyses clearly showed that M2 macrophage marker genes such as anxa1, mafb, and sepp1 were significantly decreased in WIKIM28-treated cells, whereas M1 macrophage marker genes such as hdac9, ptgs2, and socs3 were significantly increased in WIKIM28-treated PMs compared with those in WIKIM50-treated PMs.
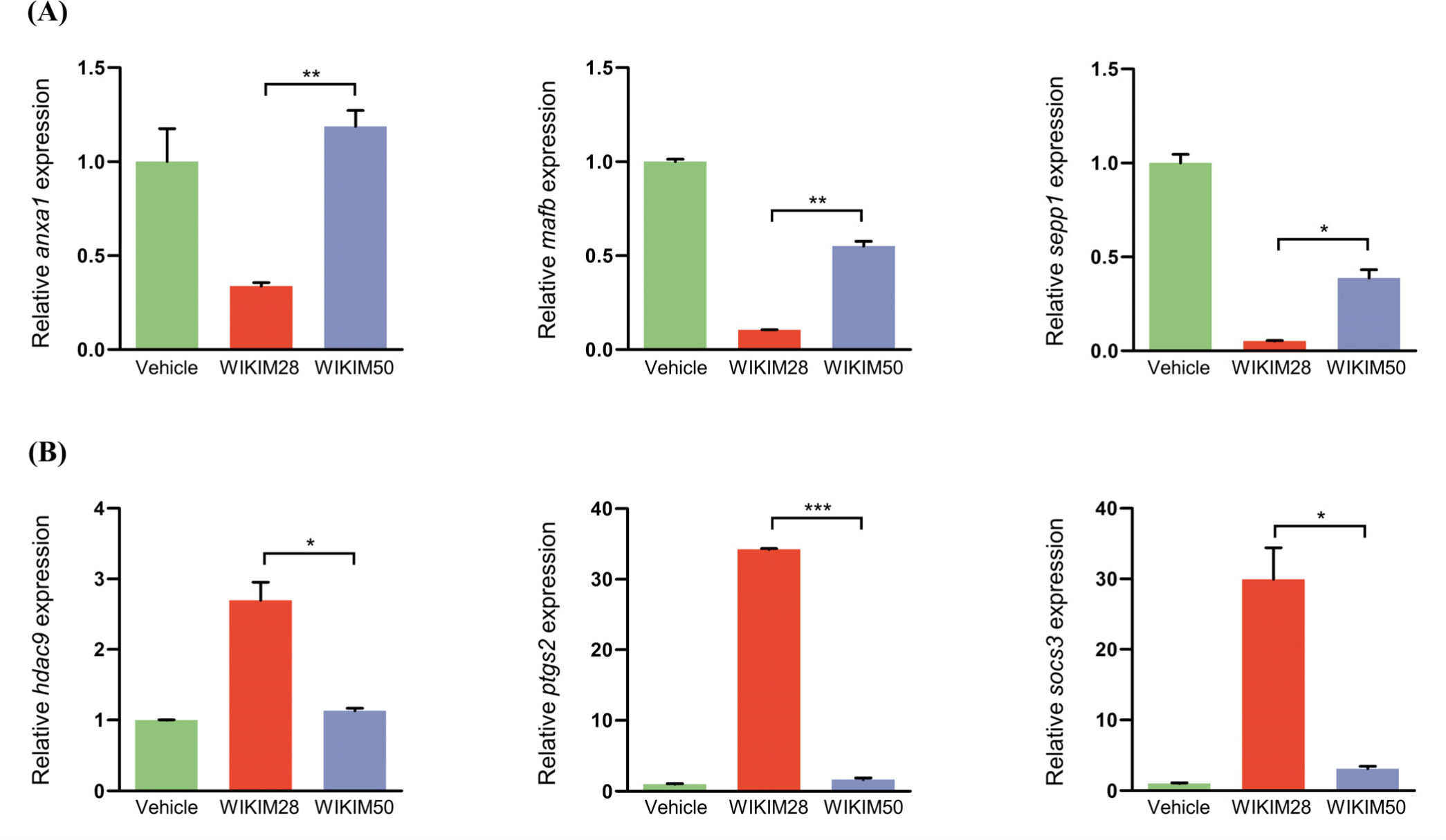
Fig. 1.
WIKIM50-treated macrophages increase mRNA transcripts of M2 signature genes and decrease mRNA transcripts of M1 signature genes compared to WIKIM28-treated macrophages.
Peritoneal macrophages (PMs; 2×106 cell/well) were co-cultured with heat killed WIKIM28 or WIKIM50 (2×107 CFU/mL) for 24 hours. Subsequently, quantitative reverse-transcriptase real-time PCRs (qRT-PCRs) were performed using total RNA from each sample as a template to determine relative mRNA expression levels of M1 or M2 macrophage signature genes. (A) Selected up-regulated genes in WIKIM50-treated PMs compared to those in WIKIM28. (B) Selected down-regulated genes in WIKIM50-treated PMs compared to those in WIKIM28. Specific gene primers of qRT-PCRs are listed in Table 1. The relative quantitation of each gene expression was normalized to expression level of mouse gapdh. All results are shown as means±SEs. * p<0.05; ** p<0.01; *** p<0.001 (n=3). Vehicle, phosphate-buffered saline (PBS)-treated macrophages; WIKIM28, WIKIM28-treated macrophages; WIKIM50, WIKIM50-treated macrophages.
Download Original Figure
Similar to our study, previous reports have demonstrated modulation of macrophage polarization by several different probiotics (Wang et al., 2020). Therefore, consumption of particular probiotics which shifts macrophage into M1 or M2 phenotype might help alleviate specific pathologic conditions of human diseases. For example, oral uptake of several different strains of probiotics can ameliorate colitis or hepatic steatosis by inducing M2 macrophage polarization in a mouse model (Jang et al., 2014; Sohn et al., 2015). However, not many studies have attempted to dissect the molecular mechanism of how certain strains can induce macrophage polarization. Here, we provide evidence indicating that signature genes of macrophage polarization are changed depending on probiotic strains. These results might be useful for finding the molecular clue of macrophage polarization caused by probiotics. Further studies will be required to define molecular signaling pathways involved in WIKIM28- or WIKIM50-mediated macrophage polarizations.
Conclusion
In conclusion, our gene chip data suggest that macrophage polarization might be modulated by treatment with different probiotic strains. Therefore, oral uptake of specific strains of probiotics might change human physiology by shifting macrophage phenotypes into either M1 or M2 polarity.
Acknowledgements
This research was supported by grants from the World Institute of Kimchi (KE1501-2 and KE2301-2).
References
Arnold CE, Whyte CS, Gordon P, Barker RN, Rees AJ, Wilson HM. 2014; A critical role for suppressor of cytokine signalling 3 in promoting M1 macrophage activation and function
in vitro and
in vivo. Immunology. 141:96-110




Barrett CW, Reddy VK, Short SP, Motley AK, Lintel MK, Bradley AM, Freeman T, Vallance J, Ning W, Parang B, Poindexter SV, Fingleton B, Chen X, Washington MK, Wilson KT, Shroyer NF, Hill KE, Burk RF, Williams CS. 2015; Selenoprotein P influences colitis-induced tumorigenesis by mediating stemness and oxidative damage. J Clin Invest. 125:2646-2660




Benjamini Y, Hochberg Y. 1995; Controlling the false discovery rate: A practical and powerful approach to multiple testing. J R Stat Soc Ser B Methodol. 57:289-300


Jang SE, Han MJ, Kim SY, Kim DH. 2014;
Lactobacillus plantarum CLP-0611 ameliorates colitis in mice by polarizing M1 to M2-like macrophages. Int Immunopharmacol. 21:186-192



Lim SK, Kwon MS, Lee J, Oh YJ, Jang JY, Lee JH, Park HW, Nam YD, Seo MJ, Roh SW, Choi HJ. 2017;
Weissella cibaria WIKIM28 ameliorates atopic dermatitis-like skin lesions by inducing tolerogenic dendritic cells and regulatory T cells in BALB/c mice. Sci Rep. 7:40040




Lu LY, Loi F, Nathan K, Lin TH, Pajarinen J, Gibon E, Nabeshima A, Cordova L, Jämsen E, Yao Z, Goodman SB. 2017; Pro-inflammatory M1 macrophages promote osteogenesis by mesenchymal stem cells via the COX-2-prostaglandin E2 pathway. J Orthop Res. 35:2378-2385




Moraes LA, Kar S, Foo SL, Gu T, Toh YQ, Ampomah PB, Sachaphibulkij K, Yap G, Zharkova O, Lukman HM, Fairhurst AM, Kumar AP, Lim LHK. 2017; Annexin- A1 enhances breast cancer growth and migration by promoting alternative macrophage polarization in the tumour microenvironment. Sci Rep. 7:17925




Shapouri-Moghaddam A, Mohammadian S, Vazini H, Taghadosi M, Esmaeili SA, Mardani F, Seifi B, Mohammadi A, Afshari JT, Sahebkar A. 2018; Macrophage plasticity, polarization, and function in health and disease. J Cell Physiol. 233:6425-6440



Sohn W, Jun DW, Lee KN, Lee HL, Lee OY, Choi HS, Yoon BC. 2015;
Lactobacillus paracasei induces M2-dominant kupffer cell polarization in a mouse model of nonalcoholic steatohepatitis. Dig Dis Sci. 60:3340-3350



Zhu XK, Yang BT, Hao ZP, Li HZ, Cong W, Kang YH. 2022; Dietary supplementation with
Weissella cibaria C-10 and
Bacillus amyloliquefaciens T-5 enhance immunity against
Aeromonas veronii infection in crucian carp (
Carassiu auratus). Microb Pathog. 167:105559

















