Introduction
From last several years, probiotics have been continuously used for health benefits for human and animals due to their beneficial effects like; tolerance to bile and acid, ability to continuously persist in gastrointestinal tract (GIT), reduction of cholesterol level, improvement of intestinal microflora, immune response stimulation (Gill et al., 2001), production of bacteriocins used as an alternatives of antibiotics (Franz et al., 2011), tumoricidal activity of natural killer cells (Matsumoto et al., 2005), capability to produce and improve the bioavailability of nutrients (Monteagudo et al., 2012; Turgis et al., 2013). Enormous amount of research in exploration and investigation is going on, to develop different and innovative nutritional supplementation approaches. In this regard, numerous health stimulating compounds such as probiotics, synbiotics prebiotics, phytobiotics and other functional food supplements have been studied (Adnan et al., 2017a). Varieties of microbial strains are currently used as probiotics in to the market. First and most commonly used are Lactobacillus and Bifidobacterium, which are indigenous to human GIT (Mombelli and Gismondo, 2000). Other newly lactic acid bacteria (LAB) includes; Leuconostoc, Pedicoccus, Lactococcus, Propionibacterium, Streptococcus and Enterococcus (Krasaekoopt et al., 2003; Power et al., 2008; Vandenplas et al., 2007; Vinderola and Reinheimer, 2003). However, it is also robustly believed that neither all probiotic do their activities with the same magnitude nor all probiotics act in the same approach. Therefore, there is an imperative need to search for newer, effective with broad range of health benefits.
Recently, marine sources have established their place in providing abundant resources for human nutrition and health. They also hold a countless varieties of living organisms having plethora of bioactive compounds with different bioactivities, when compared to terrestrial ecosystem (Adnan et al., 2018a; Hill and Fenical, 2010). Various probiotic LAB strains have been also isolated from the GI tract of different fishes (Buntin et al., 2008; Diaz et al., 2013; Ringo and Olsen, 1999), shrimps (Maeda et al., 2014), sponges (Asagabaldan et al., 2017), molluscans (Romalde and Bajra, 2010), and from other organisms (Adnan and Joshi, 2013). Many researchers during the last decade scrutinized diligently the microbial ecology of the GI tract of marine organisms (especially of fishes) (Al-Harbi and Uddin, 2004; Al-Harbi and Uddin, 2005; Hovda et al., 2007; Ringo et al., 2006a; Ringo et al., 2006b; Spanggaard et al., 2000; Yang et al., 2007; Zhou et al., 2009).
However, finding of novel strains with probiotic characteristics, which are superior to those currently in the market will satisfy the demands. Therefore, the purpose of this exploration is to isolate/validate a novel and potent probiotic strain, from the gut of freshwater fish Catla catla with a new approach and done for the first time.
Materials and Methods
Catla catla fish is commonly and freely available in market all over the world for human consumption. They were purchased from the market just for the isolation of probiotic strain from its gut. In any circumstances, live Catla catla fish was not used/harmed for any other experiments throughout this research.
Catla catla fish gut samples were enriched in MRS broth. Spread plate technique was performed onto MRS agar (Hi-Media®, India) plate with a diluted solution and incubated at 30°C for 48 h. Colonies with yellowish colour (usual for lactobacilli) were selected for further morphological examination. These colonies were stored in 20% glycerol at –20°C for future use. The putative lactobacilli isolates were first selected by confirming the catalase activity and Gram’s staining. Only Gram-positive colonies with catalase-negative activity were choosen for further scrutiny.
Identification of E. durans F3 strain was primarily carried out by using conventional biochemical methods which includes, production of acid from glucose, growth at different temperatures, NaCl concentrations, growth at different temperatures, NaCl concentrations, production of acid from glucose, catalase test homo/heterofermentative activity and Gram’s reaction. However, E. durans F3 was also additionally tested for starch hydrolysis test, phenylalanine test, IMVIC test, urea hydrolysis test, nitrate reduction test, casein hydrolysis test, triple sugar iron tests and 11 different carbohydrate fermentation (glucose, fructose, sucrose, maltose, lactose mannose, mannitol, ribose, xylose, Na gluconate, and inositol), for further identification. All the tests made it possible to identify E. durans F3 from other LAB (Harrigan and Margaret, 1976).
Further confirmation of E. durans F3 was carried out by 16S rRNA gene sequencing method. Genomic DNA was isolated from the overnight grown cells on minimal medium by NaCl-cTAB method (William et al., 2012) and quantification was done as per the method described by Sambrook et al., (1982). Later, quality of the extracted DNA (10 μL) was assessed by dissolving in Tris buffer with pH 8 (30 μL) and OD was taken at 260/280 nm. A pair of universal primer 27f (5’AGAGTTTGATCMTGGCTCAG3’) and 1492r (5’CGGTTACCTTGTTACGACTT3’) was used for carrying out the amplification of 16S rRNA gene. PCR reaction mixture contained 1x ReadyMixTM Taq PCR reaction mix (Sigma-Aldrich®, India) (10 μL), forward primer (1 μL), reverse primer (1 μL), genomic DNA template (2 μL; 50 ng/μL), and nuclease free water (6 μL). The reaction was carried out in Applied BiosystemsVeriti® thermal cycler with program adjusted as: initialization at 95°C for 4 min, denaturation at 95°C for 30 sec (35 cycles), annealing at 54°C for 30 sec, and extension at 72°C for 1 min, followed by final elongation step at 72°C for 5 min with hold at 4°C for ∞ time. Agarose gel (1%) was used for detecting the amplified PCR products with ethidium bromide as a fluorescent dye for visualizing under UV light. GenElute™ PCR Clean-up kit (Sigma- Aldrich®, India) was used for further purification of amplified PCR product. Eurofins Genomics India Pvt Ltd., Bangalore, sequenced the purified PCR product. Basic Local Alignment Search Tool (BLAST) was then used for sequence match analysis and sequences were submitted to NCBI’s GenBank database.
Clustal-W, which is a part of Molecular Evolutionary Genetics Analysis (MEGA 7.0) tool was used for carrying out the pairwise and multiple sequence alignment of 16S rRNA gene sequences of E. durans. Phylogenetic analysis was performed using the Neighbor-Joining approach in MEGA 7.0 and p-distance method was used for computing the evolutionary distances (Nei and Kumar, 2000). Taxa analyzed with evolution history was represented by bootstrap consensus tree inferred from 1,000 replicates.
E. durans F3 strain was grown in MRS broth (HiMedia®, India) for overnight at 30°C. It was sub-cultured into fresh MRS broth and incubated till the culture was grown up to 0.6 OD at 600 nm. One milliliter of culture was added into various tubes containing 10 mL of sterile MRS broth with different pH range (2.0, 3.0, and 4.0), while pH 7.0 was used as a control. After exposure to acidic conditions, viable cell counts were calculated for 0, 2, 4, and 6 h at 37°C. Survival percentage of E. durans F3 to different pH values was then calculated as percentage:
According to Huang and Adams, (2004), in vitro simulated gastrointestinal tract (GIT) conditions were prepared. Similarly, gastric juice was prepared by mixing 1 mL of sodium chloride (0.7%) (Sigma-Aldrich®, India) and pepsin (3 mg) (Sigma-Aldrich®, India), with a final pH in between 2 or 3. Whereas, simulated intestinal juice was prepared by mixing pancreatin (1 mg) (Merck®), 1 mL of sodium chloride (0.7%) (Sigma-Aldrich®, India), 1.5% of bile salts (Sigma-Aldrich®, India) with final pH 8.
E. durans F3 was grown (0.6–0.7 OD at 600 nm) into fresh MRS broth by inoculating the log phase culture, followed by centrifugation of cell culture (8,000 rpm, 10 min, 4°C). Washing of cell pellet was then performed by resuspending the cells in sterile phosphate buffered saline (PBS) (pH 7.0). Resuspended cells (0.5 mL) were added into 2.0 mL of simulated gastric or intestinal juices with incubation at 37°C for 6 h. Survival cell count were calculated at 0 (initial time) and 2, 4, and 6 h by spreading the 0.1 mL of cell suspension on MRS agar plates with incubation at 37°C. Following formula was used to calculate the survival percentage of E. durans F3 under simulated gastric juices:
According to the method of Vinderola and Reinheimer, (2003), E. durans F3 was tested for its ability to survive in the presence of bile salts. Log phase culture of E. durans F3 was inoculated (2% v/v) into MRS broth supplemented with 0.2%, 0.5%, 1.0%, 1.5%, 2%, and 2.5% (w/v) of bile salt (Hi-Media®, India) and control culture (without bile salts) with incubation at 37°C for 24 h. Percentage of growth was then calculated after OD was measured at 560 nm.
Antagonistic effects of E. durans F3 were evaluated by using agar cup/well diffusion method on Muller Hinton agar (MHA) (Hi-Media®, India) against different pathogenic organisms like P. aeruginosa, S. aureus, S. Typhi and E. coli. Test cultures were grown in a tube of nutrient broth at 37°C with adjusted turbidity to match 0.5 McFarland standards. Overnight grown culture of E. durans F3 was then centrifuged (8,000 rpm, 10 min, 4°C). Collected supernatant was then adjusted for pH 6.7–7.0 with 2 min incubation in water bath at 90°C. 0.2 mm size of syringe filter was used to filter sterilized the neutralized supernatant and inoculated into the wells made on the plates. Plates were incubated for 24 h at 37°C to observe the zone of inhibition. Sterile MRS broth was used as a negative control, while chloramphenicol antibiotic solution (100 μg/mL) was used as a positive control.
Enterocin was partially purified from overnight grown culture of E. durans F3. 100 mL of cell free extract was precipitated by the addition of ammonium sulphate (0%–90%) and centrifuged at 10,000 rpm for 10 min at 4°C. Pellet was dissolved in the PBS (pH 7.0) and dialyzed using dialysis membrane. It was then further purified through a gel filtration chromatography packed with sephadex G-100 column (Sigma-Aldrich®, India) pre-equilibrated with PBS (pH 7.0). The fraction were collected and concentrated in a lyophilizer. Sodium dodecyl sulphate polyacrylamide gel electrophoresis (SDS-PAGE) system was then used to determine the molecular weight of the purified enterocin using molecular weight marker (Merck®, India) according to the method of Laemmli (1970). SDS gel was further stained with Commassie Brilliant Blue R-250, followed by destaining i.e. overnight washing with a mixture of acetic acid-methyl alcohol-water (5:5:1 v/v).
E. durans F3 was screened for the presence of genes (entA, entB, and entP) encoding enterocins by PCR. Specific PCR primers were synthesized on the basis of published sequences for the amplification of these genes (Cintas et al., 2000). PCR was run with 1x final concentration of ReadyMixTM Taq PCR reaction mix (Sigma-Aldrich®, India) under following condition: initialization at 95°C for 4 min, denaturation at 95°C for 1 min (35 cycles), annealing at 50°C for 1 min, and extension at 74°C for 1 min, followed by final elongation step at 74°C for 5 min with hold at 4°C for infinity time. Agarose gel (1%) was used for detecting the amplified PCR products with ethidium bromide as a fluorescent dye for visualizing under UV light.
Antibiotic susceptibility test was determined semi-quantitatively using the agar overlay diffusion method of National Committee for Clinical Laboratory Standards (1993).
Antioxidant activity of cell free extract and intake cell extract of E. durans F3 was measured against α-diphenyl-α-picrylhydrazyl (DPPH) in terms of radical scavenging ability (Adnan et al., 2018b). For cell free extract, 100 μL of the supernatant, which was used earlier during antibacterial activity, was added in a tube containing 2 mL of 6×10–5 DPPH solution in ethanol (Sigma-Aldrich®, India). For intake cell extract, log phase culture was harvested by centrifugation (6,000 rpm, 10 min, 4°C). Cell pellet was then washed and resuspended in PBS (pH 7.0). 100 intact cells were separately mixed with 2 mL of 6×10–5 DPPH solution in ethanol (Sigma-Aldrich®, India), followed by 30 min incubation in dark. After the incubation period, reduction of DPPH free radicals was measured at 517 nm. One milligram per milliliter of L-ascorbic acid (Sigma-Aldrich®, India) was used as a positive control, DPPH solution (lacking sample) was used as a negative control and ethanol was used as a blank. Following formula was used to calculate the percentage inhibition of DPPH radicals:
Where, A0=absorbance of the control
A1=absorbance of the sample
Presence of the lipase enzyme which can catabolize lipid (fat & oil) from the medium was carried out on Spirit Blue Agar (SBA) (Hi-Media®, India) medium. Using sterile cork borer, 6 mm size of wells were made on the SBA plates. They were then filled with the 100 mL of fresh culture, followed by incubation for 24–48 h at 37°C. Visualization of clear blue colour halos around the wells denotes the presence of lipase.
MRS agar plates supplemented with 0.5% biles like sodium glycocholate (GCA), sodium deoxycholate (DCA) and 0.04% CaCl2 was used to assess the Bsh activity of E. durans F3 (Kumar et al, 2010). After the incubation of plates for 24 to 48 h at 37°C, presence of precipitated bile salts around the colonies with a silvery shining was observed to confirm the Bsh activity.
E. durans F3 strain was tested for pathogenicity via hemolytic and gelatinase activity. Hemolytic activity was tested on blood agar plates supplemented with 5% human blood (Hi-Media®, India). Hemolytic reaction was recorded after 24–48 h by observing the α, β and γ -hemolysis.
Gelatinase activity was determined using the fresh culture of E. durans F3, which was streaked on to the nutrient gelatin agar plate, followed by incubation for 48 h at 37°C. After the period of incubation, plates were flooded with the saturated ammonium sulphate solution. Presence of gelatinase was confirmed by a clear zone around the colonies.
E. durans F3 adhesion ability to hydrocarbon was determined by the modified method of Vinderola and Reinheimer (2003). Overnight grown culture of E. durans F3 was centrifuged (6,000 rpm, 10 min, 4°C). Cell pellet was then washed and resuspended in phosphate buffer saline (pH 7.0). Absorbance of cell pellet was adjusted to 0.6 OD at 600 nm. E. durans F3 cell suspension (2.0 mL) and xylene or n-hexadecane (1.0 mL) (Sigma-Aldrich®, India) were mixed by vortexing and incubated for 1 h at 37°C for phase separation. After incubation period, aqueous phase was taken out and absorbance was measured at 600 nm. Percentage cell hydrophobicity was calculated as percent decrease (△Abs×100) in the absorbance of the aqueous phase after mixing and phase separations relative to that of original suspension (Absinitial) as following formula:
E. durans F3 cell aggregation assay was performed by modified method of Kos et al., (2003). Supernatant was collected by centrifugating (6,000 rpm, 10 min, 4°C) the freshly grown culture of E. durans F3 (Step 1). Washing of cell pellet was then performed by resuspending in PBS (pH 7.0) and absorbance was adjusted to 0.6 OD at 600 nm (Absinitial). Suspension was then centrifuged again and the pellet was dissolved in equal volume of supernatant from step 1, followed by incubation for 2 h at 37°C. At the end of incubation, 2 mL of the upper layer was taken and absorbance (Absfinal) was measured at 600 nm by using broth as a blank. The percentage difference between initial and final absorbance as an index of cellular autoaggregation can be calculated as follows:
E. durans F3 ability to coagulate the milk was carried out against three varieties of milk obtained from the market (Whole, semi-skimmed and skimmed). 10 mL of milk were filled in all the tubes and autoclaved. Tubes were then inoculated with 1 mL of overnight grown culture of E. durans F3, followed by incubation for 24–72 h at 37°C.
Results
A LAB strain named as E. durans F3 belongs to the genus Enterococcus was isolated from the gut of Catla catla on MRS (de Man, Rogosa and Sharpe) agar plate supplemented with BCP (bromocresol purple) as pH indicator on the basis of morphological, biochemical and 16S rRNA sequencing method (Fig. 1). Colonies of E. durans F3 were appeared as small, circular with entire margin. It was facultatively anaerobic, catalase negative, able to ferment glucose, can produce ammonia from arginine, can grow at 45°C and 6.5% NaCl concentration and Gram-positive cocci in shape. However, the sugar fermentation profile and other various biochemical tests results are represented in Table 1. Nucleotide sequences were deposited to NCBI with accession numbers KF496214 after the successful identification.
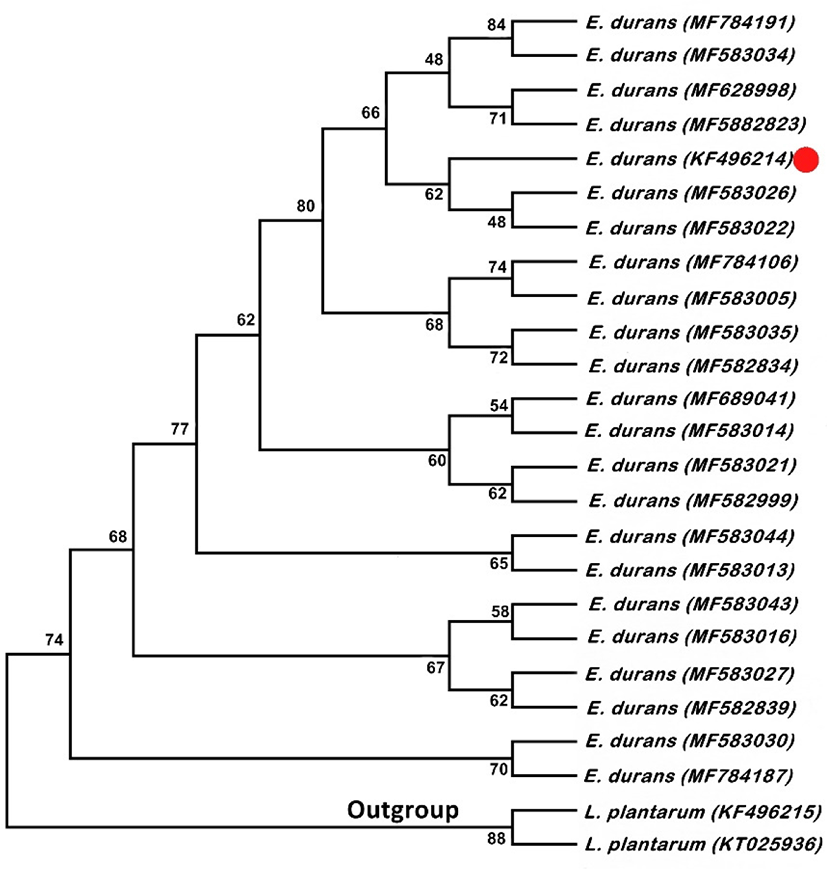
16S rRNA gene sequences of E. durans with L. plantarum as outgroup. P-distance method was used for computing the evolutionary distances. Twenty five nucleotide sequences were involved in the analysis and positions containing missing data and gaps were excluded with having the 636 positions in the final data set. Fig. 1 shows the Neighbor-Joining tree. Ratio of replicate trees is shown above the branches in which the associated taxa clustered together in the bootstrap test. Although, these results confirm that the E. durans F3 strain unequivocally belongs to the clade of genus Enterococcus.
The most anticipated characteristic necessary for probiotics is their ability to survive in the presence of bile salts under acidic conditions. E. durans F3 demonstrated high tolerance to acidic environments (Fig. 2A), excluding pH 2, where 25% of the viability was present after 2 h. No significant differences were observed at pH 3 and pH 4 when compared to control (pH 7). Incubation time was range from 0 to 6 h.
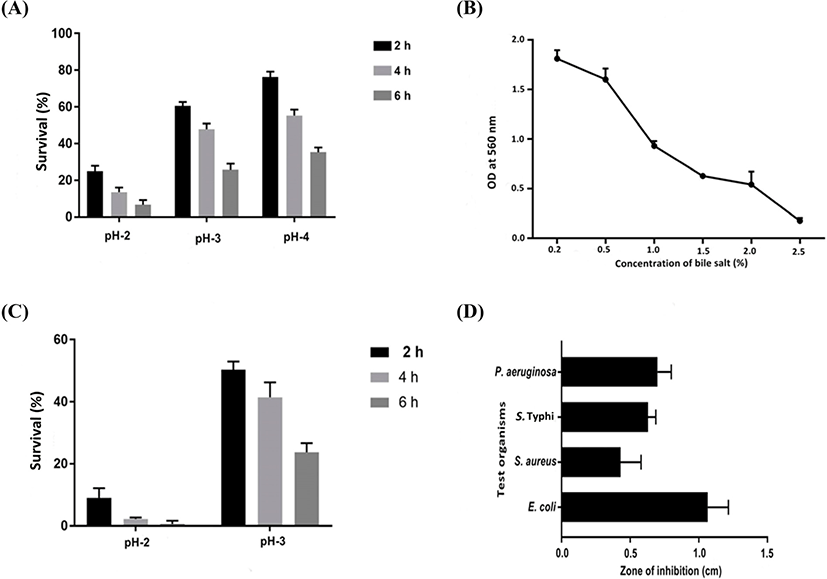
The bile tolerance of E. durans F3 was investigated from 0.2%–2.5% of bile salts (Fig. 2B). Outcome of the test revealed that E. durans F3 was able to grow up to 2.0% of bile salt concentration in a medium. However, under simulated gastric juice conditions at pH 2, E. durans F3 was survived only at time 0, and after 2 h, cell viability was not occurred (Fig. 2C). Moreover, no significant difference in cell viability was observed under simulated gastric juice condition with pH 3 when compared with control. Similar values of cell viability were observed after 6 h under simulated intestinal juice conditions and control demonstrating that E. durans F3 was able to survive under simulated intestinal juice at pH 8.
Array of pathogenic bacteria were tested against Cell free extract of E. durans F3 using the agar cup diffusion assay. In vitro tests under neutralized pH were achieved and results confirmed the inhibitory activity of E. durans F3 in the form of zone of inhibition. The strain was considered to be active, as it inhibited/supressed the growth of one or more pathogenic strains (E. coli, P. aeruginosa, S. Typhi and S. aureus) (Fig. 2D).
Purification of enterocin produced by E. durans F3 was carried out by ammonium sulphate precipitation, which was then further purified by gel filtration chromatography packed with Sephadex G-100 column. Fractions containing enterocin activity was detected by well diffusion assay. Following pre-concentration by lyophilizer, SDS-PAGE was used for testing the purity and mass of the active fraction. Protein band with an approximate molecular mass of 6.5 kDa was observed. This result suggests that the enterocin produced by E. durans F3 has an approximate molecular mass of 6.5 kDa (Fig. 3A).
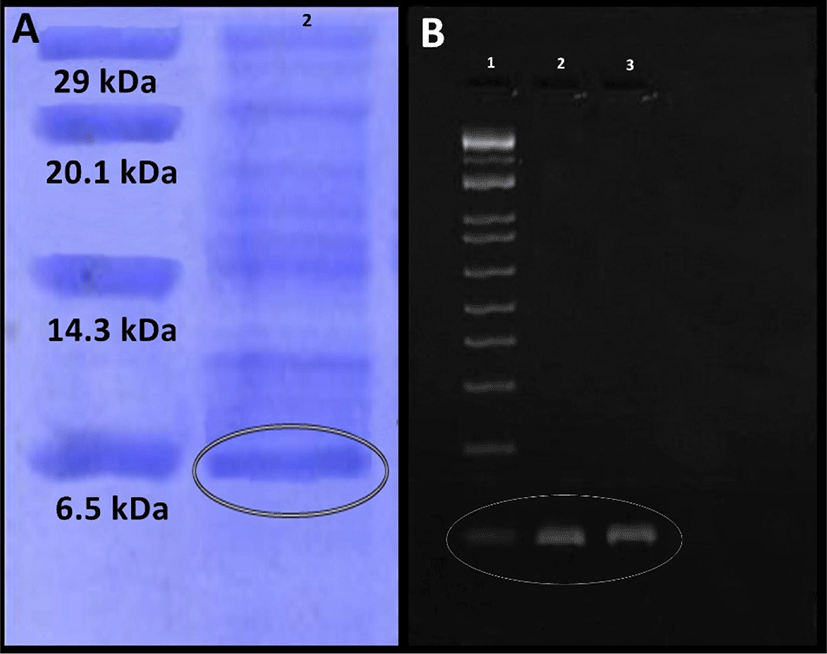
Further screening of E. durans F3 strain was done for confirming the presence of known genes which encodes class II enterocins A, B and P using PCR. Total isolated DNA from the enterocin producing E. durans F3 strain was subjected to amplification with primers to the enterocin genes. E. durans F3 showed bands corresponding to the entA (nearly at 100 bp) (Fig. 3B). No reaction took place with the enterocin B and P primers, which suggested that Bacteriocins produced by E. durans F3 are similar to enterocin A.
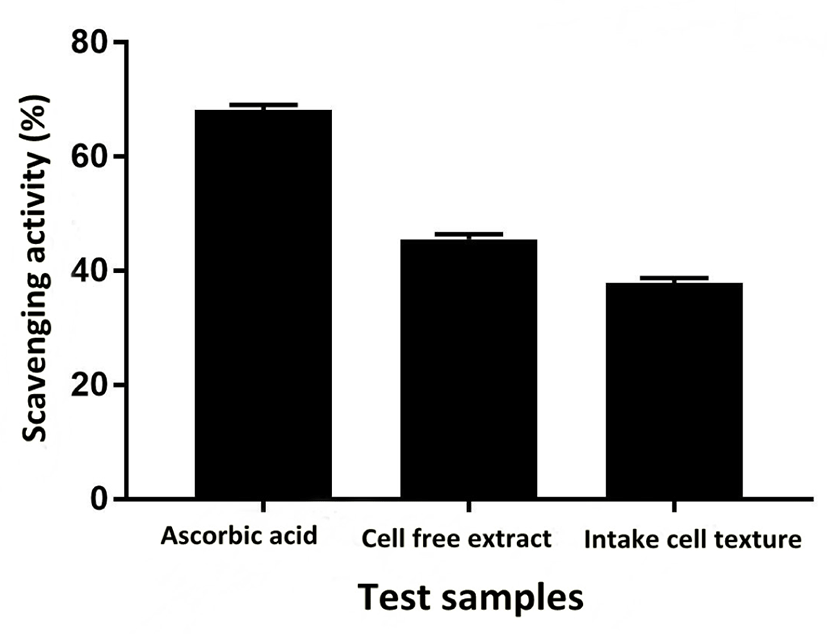
Sensitivity and resistance level of E. durans F3 was tested using antibiotic susceptibility test against different standard antibiotics. Fig. 4G shows the complete antibiotic susceptibility profile. E. durans F3 was found to be susceptible to all tested antibiotics (vancomycin, ciprofloxacin, gentamycin, linezolid, streptomycin and penicillin). The zone of inhibition around the antibiotic discs in Fig. 4G illustrates the commendable susceptibility of E. durans F3. Antioxidant potential of E. durans F3 cell free extract and intake cell extract was evaluated against α-diphenyl-α-picrylhydrazyl (DPPH) free radicals in comparison to ascorbic acid. Cell free extract was better DPPH scavengers compared to intake cell extract (Fig. 5). The higher cell free extract antioxidant activity of E. durans F3 suggests that it can assist well as a probiotic aide in scavenging free radicals.
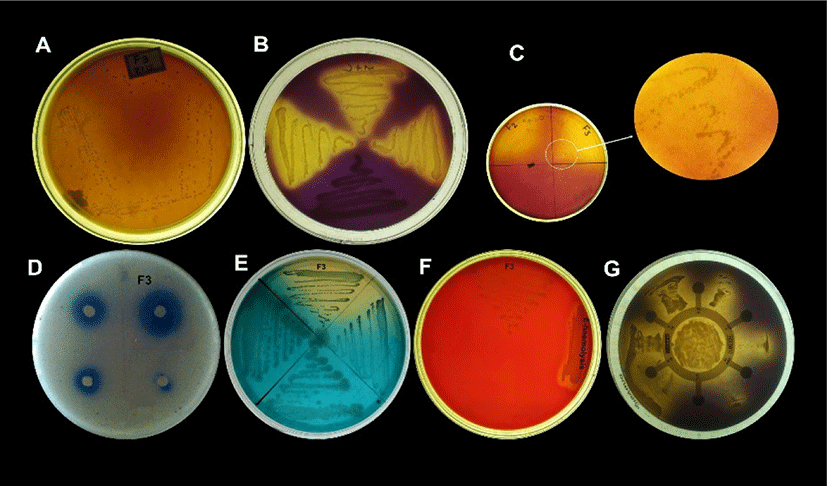
Lipolytic activity of E. durans F3 was observed on SBA medium containing emulsified lipid and spirit blue dye. Blue colour halos around the well was observed. This demonstrates the ability of E. durans F3 to produce lipolytic enzyme indicating lipolysis (Fig. 4D).
Bsh activity is also considered as one of the important factor in selection of probiotics, which found to reduce serum cholesterol. Presence of precipitated bile salts around the colony of E. durans F3 on MRS agar plate supplemented with DCA and GCA with a shine confirmed its ability to hydrolyze DCA and GCA via the production of Bsh enzyme (Fig. 4C).
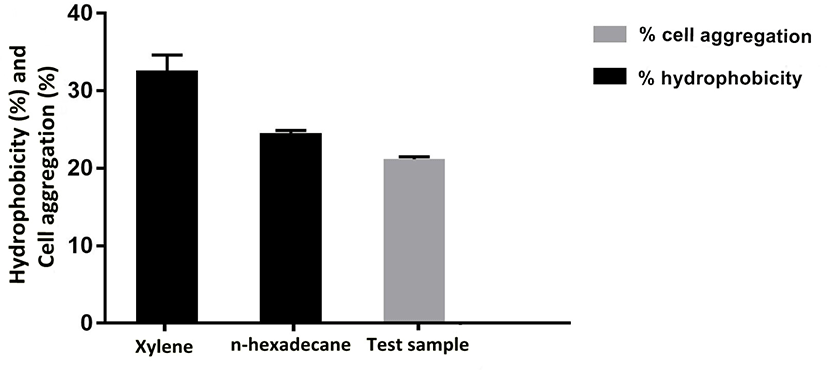
Adhesion of probiotic strains depends on cell surface properties like hydrophobicity and extracellular protein profiles, which also varies among probiotic strains. Therefore, adhesion ability of E. durans F3 was measured by cell aggregation and hydrophobicity assays using two hydrocarbons (xylene and n-hexadecane). Percent cell surface hydrophobicity of E. durans F3 was found to be 38.7% in the presence of xylene and 34.4% in case of n-hexadecane. To evaluate the cell aggregation potential of E. durans F3, the cellular auto-aggregation was measured. E. durans F3 results showed higher capability to self-aggregate (Fig. 6).
Even after 48 h of incubation on blood agar plates, E. durans F3 did not exhibited any hemolytic effect, green zone (α-hemolysis) or inhibition zone (β-hemolysis), which is considered as γ-hemolysis. However, E. durans F3 was also found to be negative for gelatinase activity. Therefore, it is considered as non-pathogenic (Fig. 4F).
Discussion
Large number of probiotic strains has been isolated mainly from the terrestrial sources successfully, with the ability to provide health benefits for a long time. Whereas, marine environment is the least explored on the planet, especially in concern of microorganisms, which are estimated to exceed 10 million species in such habitats (Grassle and Maciolek, 1992). Marine environments cover many ranges of habitats with very low to extreme high temperatures, −32°C to 400°C. Such kinds of habitats are the home of a wide diversity of microorganisms, which are acclimatized to the high temperatures, high pressure, and acidity (Gerday and Glansdorff, 2007; Merkel et al., 2013; Vetriani et al., 2004). Similarly, countless marine organisms reside in such habitat, which is believed to contain a unique microbial flora. Isolation of bacterial communities from such sources might be very helpful in identifying a potential probiotics with health benefits.
In the present study, E. durans F3 exhibited desired features to be a potent probiotic strain. Various Enterococcus species (E. faecium, E. durans, and E. faecalis) have been commonly isolated from processed foods, which might be associated to various environmental factors like extreme salinity, heat resistance and other harsh conditions (Giraffa, 2003; Martin-Platero et al., 2009). Even though, there have been studies reported on the utilization of Enterococcus species as probiotics, very few reports on their isolation, especially of E. durans from the digestive tract of aquatic animals (Sarra et al., 2013). This study is the first one, which describes the potentiality of E. durans as a potent probiotic recovered from the gut of fish with wide-ranging experimental evidences. E. durans F3 was identified on the basis of morphological, biochemical as well as 16S rRNA sequencing method.
With a vision to reveal the probable usage of the E. durans F3 as a potent probiotic, it was subjected to detailed categorisation in its probiotic profiles. In this sense, it has to survive under acidic conditions, in presence of bile salt in the GI tract. Generally, pH of the stomach ranges from 2.5 to 3.5 (Huang and Adams, 2004) and increase up to 6.5 after ingestion of food (Johnson, 1977). Thus, to remain viable under such extreme environment after oral administration, probiotic cultures faces major physiological challenges. Results in this study is proving the potentiality of E. durans F3 for tolerating acid and bile with survival under pH 3.0 and 4.0 when exposed. Even under the pH 2.0, 25% of the viability after 2 h and 2.0% concentrations of bile salts was seen. Further confirmation of bile salt tolerance was established by screening the Bsh enzyme, which can hydrolyze bile salts DCA and GCA. However, the acid tolerance ability of probiotic bacteria depends on the association of the cytoplasmic membrane and pH profile of Hþ-ATPase. Yet, it also depends on the type of bacterium, incubation conditions and organization of development of medium (Madureira et al., 2008).
After passage from the gastric barrier, probiotic bacteria have to survive the second barrier; small intestine to transit through the GI tract. Though, pH of small intestine is in favorable range but existence of bile salts and pancreatin may inhibit the growth. E. durans F3 was found to be remain viable in the presence of intestinal juice containing pancreatin at pH 8.0 and simulated gastric juice containing pepsin at pH 3.0. Thus, its survival to such simulated gastrointestinal conditions makes it as an future alternate source for probiotic based food products.
E. durans F3 also showed the ability to deter the growth of pathogens like S. aureus, S. Typhi, P. aeruginosa and E. coli. Its antibacterial activity can be said due to the production of enterocin. Enterococcus species are unique in that sense, as they can produce wide-ranging and structurally diverse enterocins; enterocins L50A, L50B (Cintas et al., 1998) and enterocin Q (Cintas et al., 2000) that are totally different from the lactic acid producing bacteriocins (Franz et al., 2007). Production of structurally diversified enterocins permits them to live in a widespread ecological niches and increase their use in food preservation (Foulquie Moreno et al., 2006). In the present study, SDS-PAGE gel was used for determining the molecular size of the enterocin produced by E. durans F3 strain. One antimicrobial peptide band with a molecular size of about 6.5 kDa was observed and verified by amplifying the expected fragment, which contains the structural gene of enterocin A.
Antioxidant activity of E. durans F3 was also observed against DPPH free radicals. Both cell free and intake cell extract of E. durans F3 were found to possess antioxidant activity. Free radicals reactive nitrogen species and reactive oxygen species are induced inside the body, which can trigger a number of human diseases via altering lipids, proteins, and DNA (Lobo et al., 2010). Hence, E. durans F3 can act as an external and additional source of antioxidants, which can contribute in controlling the oxidative stress. Performing antibiotic susceptibility test, which is an essential precondition for a strain to be a probiotic, tested further for potentiality of E. durans F3. Strains having resistance traits can impart adverse consequences to human health as they can transfer antibiotic resistance gene in to intestinal pathogens. E. durans F3 was found to be sensitive to vancomycin, ciprofloxacin, gentamycin, linezolid, streptomycin and penicillin.
Adhesion and aggregation is a step required by a potent probiotic for colonization to the intestinal epithelial cells and mucosal surface. It is likely to be a requirement for the competitive omission of enteropathogens into the gut by forming biofilms or modulating the immune response of the host (Adnan et al., 2017b; Alander et al., 1997; Forestier et al., 2001; Lee et al., 2003; Plant and Conway, 2002). Significant relationship between aggregation and cell surface hydrophobicity was shown by E. durans F3. This relationship indicating it capability of creating an antibacterial environment in the gut, by excreteing enterocin or its like substances for inhibiting the growth of pathogenic bacteria, while adhered to epithelial cells.
Evaluation of hemolytic action is also considered as one of the safety viewpoint in selecting the probiotic strains as per the guidelines from FAO/WHO. Results in this study confirmed the non-appearance of hemolytic activity. Furthermore, E. durans F3 was also able to produce lipase enzyme which helps the functioning of body in various ways; absorption of minerals and vitamins, boosting immune system, maintaining the optimum levels of pancreatic enzymes, avoiding excess weight gain and controlling obesity (Edward, 2011). Curd formation ability of E. durans F3 is also suggested in this study, due to its homofermentative nature. This makes it a impeccable starter culture for producing fermented/probiotic products imparting health benefits in dairy industries, which may also help in modifying the texture and enhancing the flavor of the final product under controlled conditions. In summary, the conclusion and outcomes of this study claims that, E. durans F3 demonstrated desirable probiotic properties in vitro.
This study emphasizes the desirable in vitro probiotic benefits of E. durans F3. Antagonistic effects of substances produced by E. durans F3 on a vast range of pathogenic bacteria indicated its important role in food preservation and human health. It can be used for the production of various kinds of novel pharmaceutical products, and functional foods. Use of E. durans F3 is not only confined to the above mentioned applications, instead it can also be used in animal farming as probiotic application in different livestock production systems like poultry, pig and ruminant nutrition. This can enhance the growth rate, feed intake, feed efficiency, carcass yield, carcass quality, nutrient digestibility and controlling of enteric pathogens. Moreover, to reduce the worldwide concern of antibiotic resistance in livestock, E. durans F3 will stand as a promising and efficient alternative. Various benefits have been observed in animal fed with different LAB-probiotics (Vieco-Saiz et al., 2019). Similarly, production of E. durans F3 originated probiotics for livestock feed possibly will fight and help in preventing bacterial diseases. It may also help in weight gain in affected animals, enhancing the quality of the products of animal farming, improve aquaculture water quality. E. durans F3 can also combine with other probiotic species, probiotics or enzymes for its use as animal feed in livestock production. In terms of future design, recombinant E. durans F3-probiotics may offer additional advantages as L. plantarum NC8, which can produce a recombinant dendritic cell-targeting peptide with activity against H9N2 avian influenza virus in chickens (Shi et al., 2016; Wang et al., 2017). Considering all of these indicated properties/activities, it can be a new promising probiotic strain.













