Introduction
Human listeriosis is one of the most common foodborne diseases in European Union (EU) and USA [European food safety authority (EFSA), 2010; Scallan et al., 2011]. In 2014, a total of 2,161 cases of listeriosis were recorded in the EU (EFSA, 2016). Also, the Center for Disease Control and Prevention (CDC) estimates that 1,600 illnesses and 260 deaths caused by listeriosis, occur annually in the United States (US) (CDC, 2016). According to a recent study, the genus Listeria consists of 17 species with validly published names. Of these, Listeria monocytogenes and L. ivanovii are considered pathogenic (Weller et al., 2015) and only L. monocytogenes are regarded as foodborne pathogens and pose a threat to public health in human (Escolar et al., 2017). In general, L. monocytogenes virulence assessment has been performed using laboratory animals such mice, guinea pigs and monkeys (Cabanes et al., 2008). However, the use of such models often involved ethical and financial burdens. Furthermore, the breeding cycle of mammals and the number of animals required became challenging factors in animal-related research. Due to these limitations, various surrogate animal models such as invertebrates including Galleria mellonella, Drosophila melanogaster, and Zebra fish (Danio rerio) have been used to evaluate host-pathogen interactions with L. monocytogenes (Chambers et al., 2012; Martinez et al., 2017; Shan et al., 2015).
Among these surrogate animal models, Caenorhabditis elegans has number of practical advantages as a model system for screening; the process is easy to perform, rapid, low-cost, can be scaled-up, and ethical acceptability (Park et al., 2014). Furthermore, the body of this nematode is transparent, allowing clear observation of all cells in mature and developing animals. Also, they have intestinal cells that are similar in structure to human intestinal cells (Irazoqui et al., 2010). By confirming that the majority of human disease genes and disease pathways are commonly present in C. elegans, the use of this nematode has emerged as promising model for the assessment of virulence of numerous human pathogens (Kaletta and Hengartner, 2006).
The main objective of this study was to perform a risk assessment of L. monocytogenes associated with the major foodborne outbreaks using C. elegans model. Furthermore, we aimed to demonstrate the importance of L. monocytogenes virulence factor by confirming the regulation of C. elegans immune genes against L. monocytogenes.
Materials and Methods
C. elegans CF512 fer-15(b26) II; fem-1(hc17) IV (fer-15; fem-1 worms) strains were used in this study. C. elegans strains were routinely maintained on nematode growth medium (NGM) plates seeded with Escherichia coli OP50 using standard procedures (Kim and Mylonakis, 2012). L. monocytogenes ScottA (serotype 4b) (Olier et al., 2002), L. monocytogenes strains 17, 18, 70, 106, 303, 3982, 3990, V7, Brie-1, LCDC (Yun et al., 2012), and L. innocua KU011 (obtained from Lab. of Food Microbiology in Korea University) were cultured on brain-heart infusion medium (BHI; BD Science, San Jose, CA, USA) at 37°C.
The C. elegans solid killing assay was performed as described previously, but with slight modifications (Kim and Mylonakis, 2012). Briefly, we prepared listeria pre-conditioning plates by spreading 20 μL of overnight cultured Listeria strains (approximately ca 2.0×109 CFU/mL) on NGM agar plates. Worms were synchronized through hypochlorite bleaching, hatched overnight and were subsequently cultured on NGM plates with E. coli OP50. Synchronized L1 larvae were transferred to NGM plates with E. coli OP50, and worms were allowed to develop to L4 larvae. On day 3, we used 30 L4/young adult hermaphrodites per plate and also transferred the worms to listeria pre-conditioning, incubated at 25°C. Worms were transferred to a new plate after every 24 h to determine the survival rates of the worms. The worms were considered dead when they did not respond to touching with a platinum wire pick. Each experimental condition was carry out with triplicate.
To measure the colonization of C. elegans, the numbers of bacterial cells in worm intestines were measured (Kim and Mylonakis, 2012). Synchronization of worm cultures was achieved by hypochlorite treatment. The synchronized L4 larvae were transferred to NGM plates with Listeria strains. After exposing C. elegans to listeria pre-conditioning plates for 24 h, 10 worms were picked randomly, washed twice in M9 buffer, and placed on BHI plates containing 100 μg/mL kanamycin (Sigma-Aldrich, St. Louis, MO, USA) and 100 μg/mL streptomycin (Sigma-Aldrich). These plates were put in 5 μL of 25 μg/mL gentamicin solution (Sigma-Aldrich) for 5 min, and results were compared with those of E. coli OP50 as a control. Afterward, worms were washed 5 times with M9 buffer, transferred into a 1.5-mL tube containing M9 buffer with 1% Triton X-100 (Sigma-Aldrich), and mechanically disrupted using a pestle (Kontes Glass Inc., Vineland, NJ, USA). These diluted worm lysates were plated and incubated at 37°C in modified LB agar for 18 h or PALCAM agar for 48 h (Oxoid Ltd., Basingstoke, UK). Each experimental condition was carried out with triplicate.
All the RNA from worms was quickly isolated following the protocol of the TRIzol reagent (Invitrogen, Carlsbad, CA, USA) and purified using the RNeasy mini kit (Qiagen, Valencia, CA, USA) including an on-column DNase digestion with RNase-free DNase (Qiagen). After RNA isolation, 50 ng of all the RNA was used for a quantitative real-time PCR (qRT-PCR) using the qPCRBIO SyGreen 1-Step Hi-ROX (PCR Bio-systems, London, UK). qRT-PCR was performed using the StepOnePlusTM instrument (Applied Biosystems, Foster City, CA, USA). Primers were designed using Primer3Input Software (v0.4.0) and were listed as shown in Table 1. Relative expression levels were calculated using the 2−ΔΔCT threshold cycle method (Livak and Schmittgen, 2001). The control gene snb-1 was used to normalize the genes’ expression data (Kim and Mylonakis, 2012).
C. elegans survival was analyzed using the Kaplan-Meier method, and the differences were determined using the log-rank test (STATA6; STATA, College Station, TX, USA). The differences in the bacterial numbers from CFU counting were determined using the Student t test. Each experiment was performed with duplicates. A p-value of 0.05 in all replicate experiments was considered statistically significant.
Results and Discussion
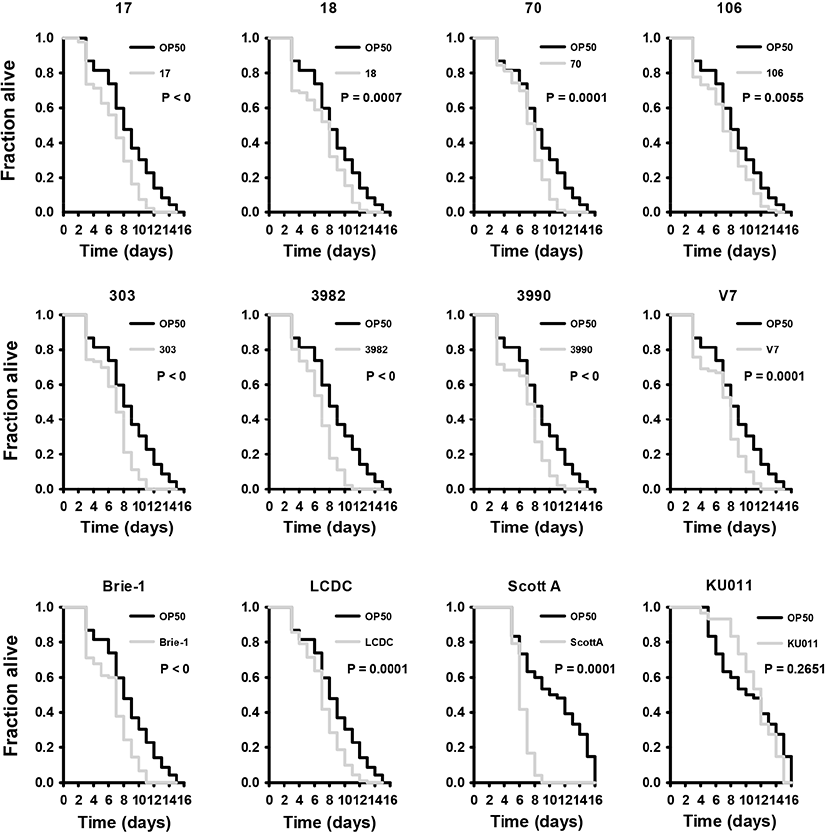
It has been well-established that C. elegans is a suitable organism for virulence models of pathogenic bacteria because they are simple and have a short lifespan as economic invertebrate animal model (Darby, 2005; Merkx-Jacques et al., 2013). In several previous studies, a relationship has been established between nematode-pathogenic bacteria and bacteria that are capable of killing mice (Sifri et al., 2002). In this study, we studied the feasibility of C. elegans for the virulence study of L. monocytogenes. Initially, we examined whether L. monocytogenes Scott A and L. monocytogenes EGD-e cause food poisoning and influences the lifespan of C. elegans. As same manner, we employed the L. innocua KU011, which is nonpathogenic listeria strain as control. The nematodes was highly susceptible when exposed to L. monocytogenes. All L. monocytogenes strains used in this study showed that C. elegans leads to faster death than E. coli OP50; however, L. innocua KU011 showed similar death period to E. coli OP50 (Fig. 1). The killing effect of C. elegans infected with L. monocytogenes were briefly reported in the previous studies (Thomsen et al., 2006). We found that C. elegans exposed to L. innocua KU011 was not significantly different from C. elegans exposed to non-pathogenic E. coli OP50 used as a standard normal feed source for worms in killing assays with fer-15; fem-1 worms (p=0.2651); hence, we concluded that both of them are not harmful to C. elegans. In addition, we found that L. monocytogenes strains also killed C. elegans much faster as compared to the avirulent E. coli OP50 and L. innocua KU011 (p<0.0001). As a result, L. monocytogenes is highly toxic to C. elegans host compared to worms feeding on E. coli OP50 and L. innocua KU011.
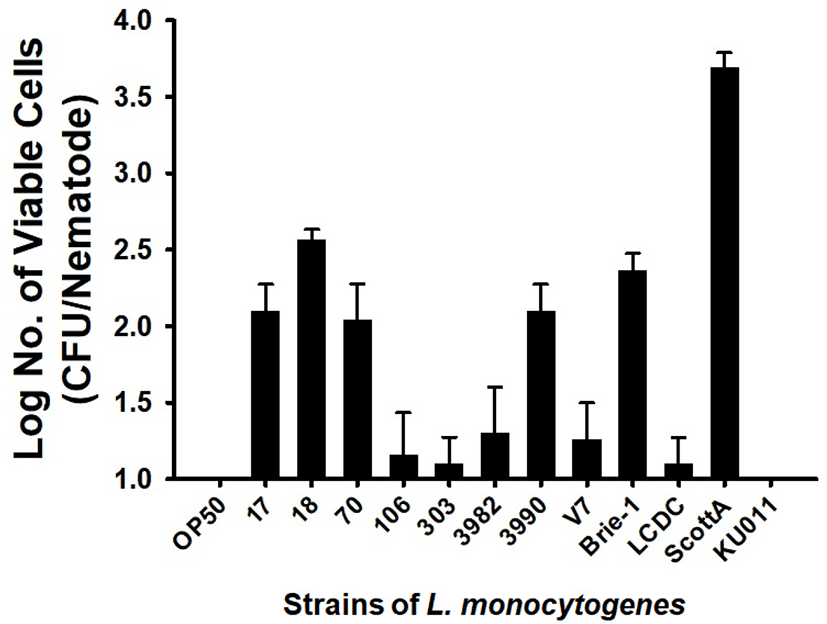
Since some pathogenic bacteria caused a persistent lethal infection in intestinal tract, we evaluated the possibility that L. monocytogenes cells colonize the C. elegans intestine, thereby killing the C. elegans. E. coli OP50 and L. innocua KU011 showed similar nematode survival rates and intestinal bacterial load. As a result, no bacterial cells could be detected in the nematode intestine 24 h after feeding on E. coli OP50 and L. innocua KU011. On the other hand, L. monocytogenes strains colonize the C. elegans intestine more than E. coli OP50 or L. innocua KU011. The strains of L. monocytogenes that were harmful to C. elegans showed high colonization of 1.10±0.17–3.69±0.10 CFU/mL in nematode intestinal tract after 24 h of exposure (Fig. 2; p<0.05 for all strains of L. monocytogenes, compared with worms feeding on E. coli OP50 or L. innocua KU011). In particular, the most toxic L. monocytogenes Scott A showed a high colony counts of 3.69±0.10 CFU/mL in the nematode intestinal tract. After being digested by the nematode, burden of Gram-positive pathogens including Staphylococcus aureus and Enterococcus faecalis in the nematode intestine are prerequisite for virulence of pathogens (Garsin et al., 2001). Thus, we concluded that L. monocytogenes strains may be pathogenic by colonizing the intestinal tract of C. elegans.
Having established that C. elegans nematodes are kill by L. monocytogenes strains infection. We aimed to determine the immune mechanism associated with infection of C. elegans with L. monocytogenes. Four strains with the highest toxicity among L. monocytogenes strains were tested. We selected 4 immunity-related target genes that critically increased by Gram-positive pathogens such as E. faecalis and S. aureus as follows; C15C8.3 (encodes aspartyl proteases), cpr-5 (encode cysteine proteases), lys-5 (encodes a lysozyme), and clec-60 (encode C-type lectins). The transcription of the nematode immune gene to L. monocytogenes strains exposure was analyzed in comparison with E. coli OP50, the standard normal feed source for C. elegans. With reference to our previous reports (Kim and Mylonakis, 2012), all of the immune associated genes were significantly upregulated by the exposure of L. monocytogenes strains (p<0.05 for all transcriptions of immune related genes in C. elegans with infection of L. monocytogenes strains, comparing with E. coli OP50). More specifically, our qRT-PCR results indicated that infection with L. monocytogenes ScottA intensely induced the transcription of C15C8.3 (6.8±0.1-fold), cpr-5 (3.3±0.4-fold), lys-5 (10.0±1.6-fold), and clec-60 (4.1±0.2-fold) (Fig. 3). L. monocytogenes strain 17, induced the transcription of C15C8.3 (11.7±1.5-fold), cpr-5 (1.6±0.7-fold), lys-5 (19.5±6.7-fold), and clec-60 (18.0±5.9-fold). For L. monocytogenes strain 303, induced the transcription of C15C8.3 (9.4±3.2-fold), cpr-5 (2.2±1.1-fold), lys-5 (19.5±3.0-fold), and clec-60 (12.4±0.4-fold). L. monocytogenes strain 3982, induced the transcription of C15C8.3 (9.2±3.5-fold), cpr-5 (4.1±1.8-fold), lys-5 (18.7±2.8-fold), and clec-60 (10.9±2.8-fold) (Fig. 3).
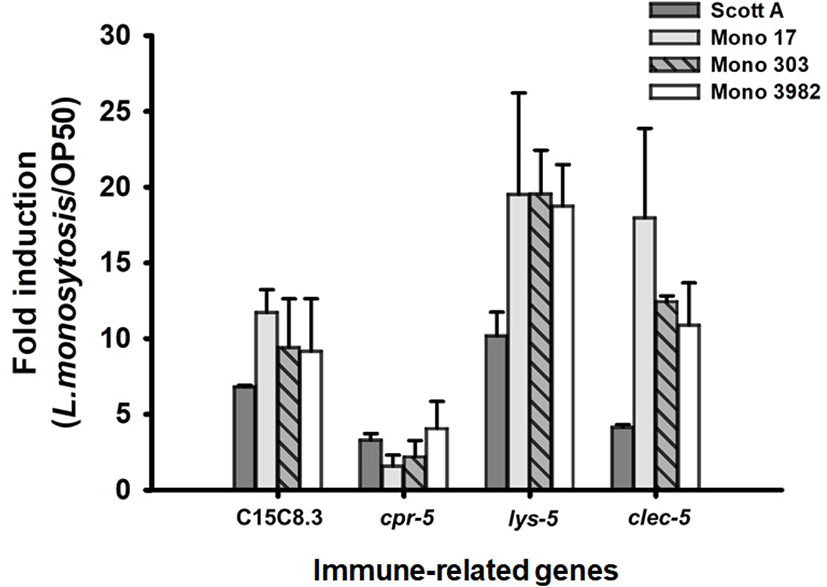
C15C8.3 encodes aspartyl proteases while cpr-5 encodes cysteine proteases. Its primary function is lumen hydrolysis of bacterial proteins (McGhee et al., 2007). Transcriptional upregulation of these genes was observed in previous studies in response to Gram-positive bacteria (Kim and Mylonakis, 2012). The gene encoding proteases were also up-regulated in gram-positive bacteria, L. monocytogenes infection. Lysozyme plays an important role in animal immunity by catalyzing the cleavage of peptidoglycan, a major component of the bacterial cell wall (O’Rourke et al., 2006). The destruction of bacterial cells is believed to play an important role both in defending against infectious bacteria and in digesting bacteria as a nutrient source (Schulenburg and Boehnisch, 2008). The lysozymes quantified in this study showed changes similar to qPCR results of a previous studies (Boehnisch et al., 2011; Treitz et al., 2015), with lys-5 increased. C-type lectin-like domain containing proteins (CTLD) have been described to play a role in binding of specific pathogen-derived molecules (Ghazarian et al., 2011). Transcriptional upregulation of genes coding for C-type lectins has been observed in previous studies in response to gram-positive bacteria (O’Rourke et al., 2006; Treitz et al., 2015). The transcription of clec-60 was also observed to have significantly increased in response to the L. monocytogenes in our study. Taken together, the induction of these genes was strongly associated with pathogenesis of L. monocytogenes. Interestingly, C15C8.3, lys-5, and clec-5 were highly expressed in the remaining strains than L. monocytogenes ScottA, which has the highest toxicity with the fastest killing of C. elegans. This results suggests that the death of C. elegans is associated with a high colony for the intestines.
In conclusion, this study established that L. monocytogenes are not digested by the nematode and they can successfully colonize in intestinal tract of nematodes. This persistence resulted in toxic condition for C. elegans host. In addition, these results indicate that conditioning with the L. monocytogenes specifically stimulated the transcription of genes associated with nematode immune response to Gram-positive pathogens.













