Introduction
Bromodomain-containing protein 2 (BRD2) is the founding member of the bromodomain and extraterminal domain family of double bromodomain-containing genes, which have two tandem bromodomains at the N-terminal region and an extra-terminal domain at the C-terminal side of the bromodomains (Shang et al., 2011). BRD2 is a signal transducer (Denis and Green, 1996) and is ubiquitously expressed in mammalian tissues (Thorpe and Beck, 1998). A BRD2 hypomorphic allele induced extreme obesity in mice fed a regular diet, but the mice did not develop insulin resistance or diabetic diseases (Wang et al., 2009). Moreover, BRD2 is required for the inflammatory process (Belkina et al., 2013) and regulates the nuclear factor-E2-related factor 2-dependent transcription of antioxidant genes (Michaeloudes et al., 2014). However, its function has not yet been studied in pigs.
Meat quality has become a primary focus of pig production. Meat quality traits in pigs tend to be moderately to highly heritable (Devine and Dikeman, 2014). When applied in breeding programs, genetic markers are useful for selecting individuals, and the higher the heritability of a trait, the more efficient and effective early phenotypic selection can be (Szabó et al., 2011). In addition, in most cases, traits of interest can only be measured in carcasses. This is difficult and expensive, and the animals being measured are no longer available for breeding. Recently, various genomics tools have been applied to overcome these limitations (Baby et al., 2014; Casiro et al., 2017; Hwang et al., 2017; Kwon et al., 2016). A candidate gene approach was successfully applied to identify several DNA markers (Van der Steen et al., 2005). Detection of single nucleotide polymorphisms (SNPs) within genes affecting meat quality traits and their associations, linkage analysis, and gene expression are the most important and commonly used tools for characterizing candidate genes. Therefore, we identified SNPs of the BRD2 gene and investigated BRD2 in detail to assess its candidacy as a marker for meat quality traits in Berkshire pigs.
Materials and Methods
Phenotypic data were obtained from 384 pigs of a pure Berkshire line. All of the pigs were reared at the same pig farm (Dasan Pig Breeding Co., Namwon, Korea) and were slaughtered at a commercial abattoir at body weights of approximately 110 kg. After slaughter, the hot carcass weight was recorded, and backfat thickness was measured between the 10th and 11th ribs. Meat quality traits were evaluated in the longissimus dorsi muscle. Thirteen meat characteristics were assessed: meat colors (L* [lightness], a* [redness], and b* [yellowness]), chemical composition (moisture, collagen, crude protein, and crude fat %), drip loss (%), cooking loss (%), water-holding capacity (%), Warner-Bratzler shear force (kg/cm2), carcass temperatures (℃) at 1, 4, 12 and 24 h after slaughter (T1, T4, T12, and T24, respectively), and the 24 h postmortem pH (pH24). Meat colors were recorded by a Minolta Chroma Meter (CR-400; Minolta, Osaka, Japan) after 30 min of blooming at 1℃. The moisture, crude protein, and crude fat contents were measured using the methods set out by the Association of Official Agricultural Chemists in longissimus dorsi samples taken 24 h after slaughter (Horwitz, 2000). Collagen content was calculated using a colorimetric assay employing a hydroxyproline standard solution. Drip loss was calculated as the weight difference before and after storage. A slice 2 cm in thickness (weight 100±5 g) taken from the longissimusdorsi was placed in a polypropylene bag (Dongbang Co., Gimpo, Korea), vacuum-packed, and stored for 24 h at 4℃. Cooking loss was calculated as the weight difference before and after heating. A slice of 3 cm in thickness (weight 100±5 g) from the longissimusdorsi was placed into a polypropylene bag (Dongbang Co.), cooked for 40 min at 70℃ in a water bath, while the internal meat temperature reached 60℃. After boiling, the bag was removed the water bath and cooled to room temperature. Water-holding capacity was measured at 3 days postmortem using a centrifugation method. Duplicate 10 g minced samples taken from one chop from each loin were placed into centrifuge tubes and spun for 10 min at 40,000×g. After centrifugation, the liquid was removed and the meat was re-weighed. The percentage of water loss was calculated and used to estimate the water-holding capacity (Fan et al., 2010). Carcass temperature was measured in the longissimus dorsi at 1, 4, 12 and 24 h post-mortem using a deep carcass thermometer (Flash Link electronic logger, Model 20209, DeltaTrack, Inc., Pleasanton, CA, USA). The postmortem pH24 was measured with the aid of a portable pH meter (Istek Inc., Seoul, Korea) equipped with a glass electrode that can be inserted into muscle tissue. Experimental protocols for this study were undertaken following the guidelines established by the Animal Care and Use Committee of GNTECH and the Korean Animal Protection Act and related laws (Permit Number: 2015-5).
Reverse transcription polymerase chain reaction (RT-PCR) was performed to determine the expression profile of the BRD2 gene in various tissues of Berkshire pigs. Total RNAs from various tissues (liver, stomach, lungs, kidney, large and small intestines, spleen, muscle, and adipose tissue) of three Berkshire pigs were isolated using TRIzol Reagent (Molecular Research Center, Cincinnati, OH, USA), and cDNA was synthesized by RT-PCR using 2 μg total RNA and SuperScript II reverse transcriptase (Invitrogen, Carlsbad, CA, USA) according to the manufacturer’s protocol. BRD2 gene (GenBank accession no. EU402599) specific primers (Forward: 5′-GCACAAACGCTGGAAAAGAT-3′ and Backward: 5′-TAGGTATCTCGGGTGGTGGA-3′) for RT-PCR were designed using Primer3 software (Rozen and Skaletsky, 2000). Amplification reactions were performed on a Perkin Elmer 9700 system (Applied Biosystems, Waltham, MA, USA) using the following conditions: 95℃ for 5 min; 30 cycles of 95℃ for 30 s, 60℃ for 30 s, and 72℃ for 30 s; and a final elongation for 7 min at 72℃. The PCR products were separated on 2% (w/v) agarose gels and quantified using a Gel Logic model 200 imaging system (Kodak, Rochester, NY, USA). The results were reported as the relative expression level after normalization of the transcript level to that of peptidylprolyl isomerase A (PPIA; GenBank accession no. NM_214353.1) (Forward: 5′-CACAAACGGTTCCCAGTTTT-3′ and Backward: 5′-TGTCCACAGTCAGCAATGGT-3′).
To identify polymorphic variants, total RNA was isolated from liver tissues in three pigs using TRIzol reagent (Molecular Research Center) according to the manufacturer’s instructions and mRNA was purified with an RNA-Sequencing (RNA-Seq) sample preparation kit (Illumina, Inc., San Diego, CA, USA) (Jung et al., 2012). RNA-Seq analyses were performed on the synthesized cDNA using an Illumina GAII analyzer (Illumina, Inc., USA). SNPs were detected by assembling and mapping raw data to UniGene, and information on the detected SNPs was obtained using the NCBI database. Genomic DNA was isolated from whole blood cells in 384 pigs and subjected to SNP genotyping analysis using the Illumina VeraCode GoldenGate Assay kit (Illumina, Inc., USA). Oligonucleotide information for the genotyping of SNPs is summarized in Table 1.
The general linear model procedure was used to analyze the association between genotypes and meat quality traits using SAS software version 9.1.3 (SAS Institute Inc., Cary, NC, USA). SNPs for statistical analysis were selected by a call rate<0.90, a minor allele frequency (MAF)>0.01, and Hardy-Weinberg equilibrium (HWE) p>0.05. The linear model was yijk=μ+Gi+Sj+Pk+eijk, where yijk was the phenotypic value of the target trait, μ was the general mean, Gi was the fixed effects of genotype i, Sj was the fixed effects of sex j, Pk was the random effects of polygenicity k and eijk was the random error. The associations were assessed using three genetic comparison models: the dominant model (the combined variant homozygote and heterozygote versus the wild-type homozygote), the recessive model (the variant homozygote versus the combined heterozygote and wild-type homozygote), and the co-dominant model (the wild-type homozygote versus heterozygote versus the variant homozygote). To verify significant differences (p<0.05) among the genotypic frequencies of traits, analysis of variance with Bonferroni correction and Kruskal-Wallis tests were used.
Results and Discussion
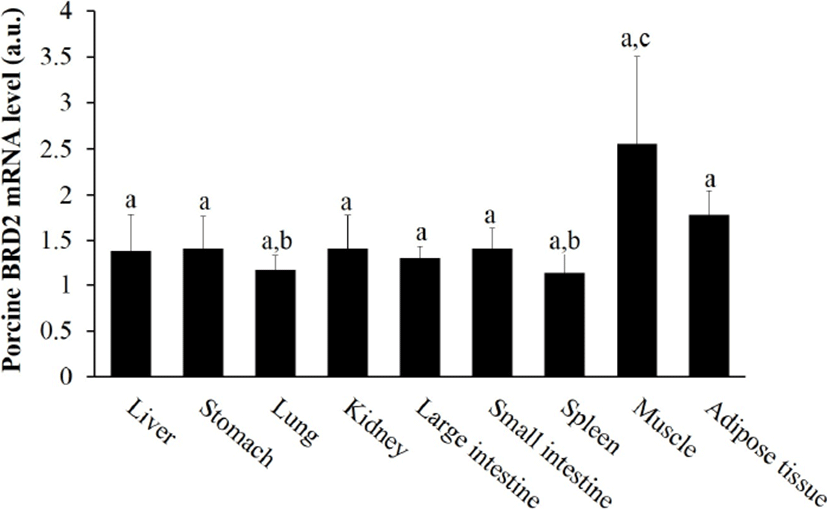
First, we examined the tissue expression profile of BRD2 mRNA by performing RT-PCR. Porcine BRD2 was ubiquitously expressed in the liver, stomach, lung, kidney, large and small intestine, spleen, muscle, and adipose tissues, similar to murine BRD2 (Taniguchi et al., 1998), but its expression was highest in muscle, followed by adipose tissue (Fig. 1). Meat quality depends on physiological processes in muscle tissue, potentially involving many genes associated with muscle structure and metabolism. We assumed that BRD2 status would be a determinant of meat quality.
Via RNA-Seq, a total of 18,969 SNPs were identified in 4,248 genes. Of them, approximately 600 SNPs which were available the location information in the gene sequence and non-synonymous, were preferentially selected. We further subjected to association analysis between the SNPs and meat quality traits in Berkshire pigs and then 96 SNPs were selected among them (Supplementary Table 1). An SNP c.1709G>C (rs196956513) in the BRD2 gene was finally selected based on the high possibility of having an effect on the meat quality. This is a non-synonymous SNP (nsSNP), with glycine changed to alanine at position 570. To analyze the genotypic and allelic frequencies of this nsSNP, 384 Berkshire pigs were genotyped using the GoldenGate assay and the results are given in Table 2. The genotype counts of the BRD2 nsSNP were 257, 101, and 13 for GG, GC, and CC genotypes, respectively. The frequency of the G allele was 0.829 and of the C allele was 0.171. The animals were in HWE (p>0.05) at the BRD2 c.1709G>C locus (Falconer and Mackay, 1996).
| Genotype | Genotype frequency | Allele | Allele frequency | MAF | HWE | Call rate |
|---|---|---|---|---|---|---|
| GG (n=257) | 0.687 | G | 0.829 | |||
| GC (n=114) | 0.284 | C | 0.171 | 0.1712 | 0.4602 | 0.9849 |
| CC (n=13) | 0.029 |
The association between the nsSNP in porcine BRD2 and meat quality traits is shown in Table 3. This nsSNP was significantly associated with a carcass weight, meat color (a* value and b* value), protein content, and postmortem pH24 in a dominant model (GG versus GC+CC). Also, significant associations between the nsSNP and a carcass weight, meat color (a* value), cooking loss, protein content, water holding capacity, carcass temperatures (T4, T12, and T24) and postmortem pH24 were observed in a codominant model (GG versus GC, GC versus CC, and GG versus CC). However, the nsSNP in the BRD2 gene was only significantly associated with a water holding capacity, carcass temperatures (T4, T12, and T24), and postmortem pH24 in a recessive model (GG+GC versus CC). Of these traits, postmortem pH24 was significantly affected by all of the models. These results suggest that the CC genotype of this nsSNP was more associated with a meat quality than the GG and GC genotypes. pH and temperature conditions are important postmortem factors known to affect meat quality, particularly drip loss and water holding capacity. In addition, the rapidly cooled carcasses resulted in a lower rate of the pH fall (Maribo et al., 1998). The lower rate of pH fall in the muscle is reduced by time from exsanguination until ultimate pH is reached, due to a reduction in the glycogen content in the muscle, a falling temperature and falling pH reduce the metabolic/enzymatic activity (Bourne, 1973).
Human BRD2 protein is a nuclear Ser/Thr kinase whose activity is increased upon cellular proliferation and is remarkably elevated in peripheral blood lymphocytes collected from acute and chronic lymphoma patients (Denis and Green, 1996). Although kinase activity has not been observed in porcine BRD2, it has more than 98% amino acid sequence identity with human BRD2. Another important function of BRD2 is to specifically recognize acetylated histones through their bromodomains to promote transcription of genes required for determining cell identities (Kanno et al., 2004). In other words, BRD2 protein fulfills a key role a “reader” of the histone code to induce transcriptional activation of various genes. Recent studies have reported that BRD2-disrupted mice showed significantly elevated levels of proinflammatory cytokines such as interleukin-1β and tumor necrosis factor-α in the blood, and induced insulin resistance and obesity (Wang et al., 2009). The BRD2-deficient animal model for “metabolically healthy” obesity in particular has received attention in an effort to understand the signal transduction pathways at work in obesity (Wang et al., 2013). In other words, as a bromodomain and external domain family transcriptional regulator, BRD2 regulates the expression of many genes by interpreting chromatin codes, and participates in the regulation of body energy balance.
Pigs need to mobilize body reserves, mainly fat, as an energy source. Body energy balance is regulated by a complex network of signals including transcriptional co-regulators such as PCG-1, a peroxisome proliferator-activated receptor gamma (PPAR-γ) co-activator which controls energy storage and expenditure by regulating the development and function of adipose tissue, skeletal muscle and liver (Handschin and Spiegelman, 2006). In 3T3-L1 pre-adipocytes, BRD2 normally co-represses PPAR-γ and inhibits adipogenesis (Zang et al., 2013). Thus, BRD2 may affect meat quality by regulating energy balance.
In conclusions, we found that the nsSNP (c.1709G>C) of BRD2 gene, which is relatively highly expressed in muscle tissue, was significantly associated with meat quality in Berkshire pigs. The quality of meat with the BRD2 CC genotype was superior to that with the BRD2 GG or GC genotypes. These findings provide information for genetic characterization or association studies in other populations. In addition, this nsSNP may be used a genetic marker for improving meat quality traits in pigs. However, further studies on different larger-scale population of pigs are needed to establish the reliability of this SNP.
Supplementary Materials
Supplementary materials are only available online from: https://doi.org/10.5851/kosfa.2018.e7.













