Introduction
The genus Weissella, which was proposed in 1993, includes species previously classified as Leuconostoc and Lactobacillus (Collins et al., 1993). Currently, there are 21 valid Weissella spp., and strains in this genus have recently received attention for their potential use as probiotics (Fessard and Remize, 2017; Fusco et al., 2015). Weissella spp. are widely found in diverse environments as well as in various fermented foods (dairy fermented products, fermented meat and fish based products, cereal based fermented food and vegetables and fruits fermented foods). They are heterofermentative lactic acid bacteria, producing CO2 from carbohydrate metabolism and lactic and acetic acids as the major end products of glucose metabolism (Björkroth et al., 2014). Weissella cibaria is a good starter for fermentation as it produces less lactic acid than homofermentative lactic acid bacteria, and some W. cibaria strains have been shown to produce high levels of exopolysaccharides, which have desirable texturizing properties (Carr et al., 2002; Fessard and Remize, 2017). Phages that infect lactic acid bacteria are a concern in the fermented food industry, especially in the production of fermented dairy foods, because phage contamination can reduce product quality (Garneau and Moineau, 2011; Kot et al., 2014). Phages are viruses that only infect bacteria, and they are the most abundant microorganisms on the earth; thus, they are widespread and are found on many foods (Hendrix, 2003). However, phages are also used in various biotechnology applications, including phage display, bacterial detection, biofilm degradation, and pathogen biocontrol (Greer, 2005; Hudson et al., 2005). Due to the host specificity of phages, they are ideal agents for controlling bacterial contamination of foods. Phage-based applications, such as phage therapy (Sulakvelidze et al., 2001), have been used to control pathogens in livestock, for sanitation, specifically the decontamination of carcasses and other raw products (Martinez et al., 2008; Modi et al., 2001), and for the control of lactic acid bacteria in the fermentation industry (Liu et al., 2015). In this study, we report the morphogenetic and genome sequence analyses of a WCP30 phage, which was isolated from a fermented food and may be useful for controlling the fermentation in dairy products.
Materials and Methods
All reagents and media were purchased from Sigma-Aldrich (USA) and BD BBL (USA), respectively. To isolate phage that infect W. cibaria, various fermented food samples were collected from a local market in Korea. W. cibaria KCTC3807 was used as the host strain and was grown in Lactobacilli MRS broth or agar supplemented with 10 mM CaCl2 (LBC) at 37°C overnight. To isolate W. cibaria phage, the samples were analyzed by plaque assay with the aforementioned host strain. Briefly, sample (5 g) was mixed with W. cibaria (7-8 Log CFU/mL) and incubated at 37°C for 24 h. Then, the culture was centrifuged, and the supernatant was filtered through a 0.22 μm membrane syringe filter. The filtrate was tested for the presence of W. cibaria-infecting phage by a plaque assay using a double agar overlay. A plaque was picked, and the phage was eluted with SM buffer (100 mM NaCl, 8 mM MgSO4•7H2O, and 50 mM Tris-Cl [pH 7.5]), and a plaque was picked from a fresh plate for plaque purification (Sambrook and Russel, 2001).
To confirm the morphological characteristics, purified phage particles were negatively stained with 2% aqueous uranyl acetate (pH 4.5) on a carbon-coated grid and examined by transmission electron microscopy. Phage DNA was collected from polyethylene glycol precipitated phage particles by method (Manfioletti and Schneider, 1988) with some modifications. DNase I (10 μg/mL) and RNase A (20 μg/mL) were added to phage lysate, respectively. After incubation at room temperature for 15 min, 0.5 M EDTA (pH 8) and proteinase K (1 mg/mL) were added, followed by incubation at 65°C for 30 min. After incubation, the nucleic acid was extracted with phenol-chloroform-isoamyl alcohol. The nucleic acid was precipitated with ethanol, and resuspended in sterile distilled water. Phage DNA was stored at −80°C.
The genome sequence was determined by ultra-high throughput GS FLX sequencing (average coverage, 20-fold redundancy). Then, the obtained nucleotide sequence was compared to sequences in GenBank by BLAST (http://www.ncbi.nlm.nih.gov/BLAST/). The open reading frames (ORFs) were identified with ORF Finder at the National Center for Bioinformatics (http://www.ncbi.nlm.nih.gov/gorf.html). The molecular weights and isoelectric points of the predicted proteins were calculated with the Compute pI/Mw program (http://www.expasy.ch/tools/pi_tool.html). The promoters and tRNAs were identified with promoter hunter (http://www.phisite.org) and tRNAscan-SE (http://lowelab.ucsc.edu/tRNAscan-SE/), respectively.
Results and Discussion
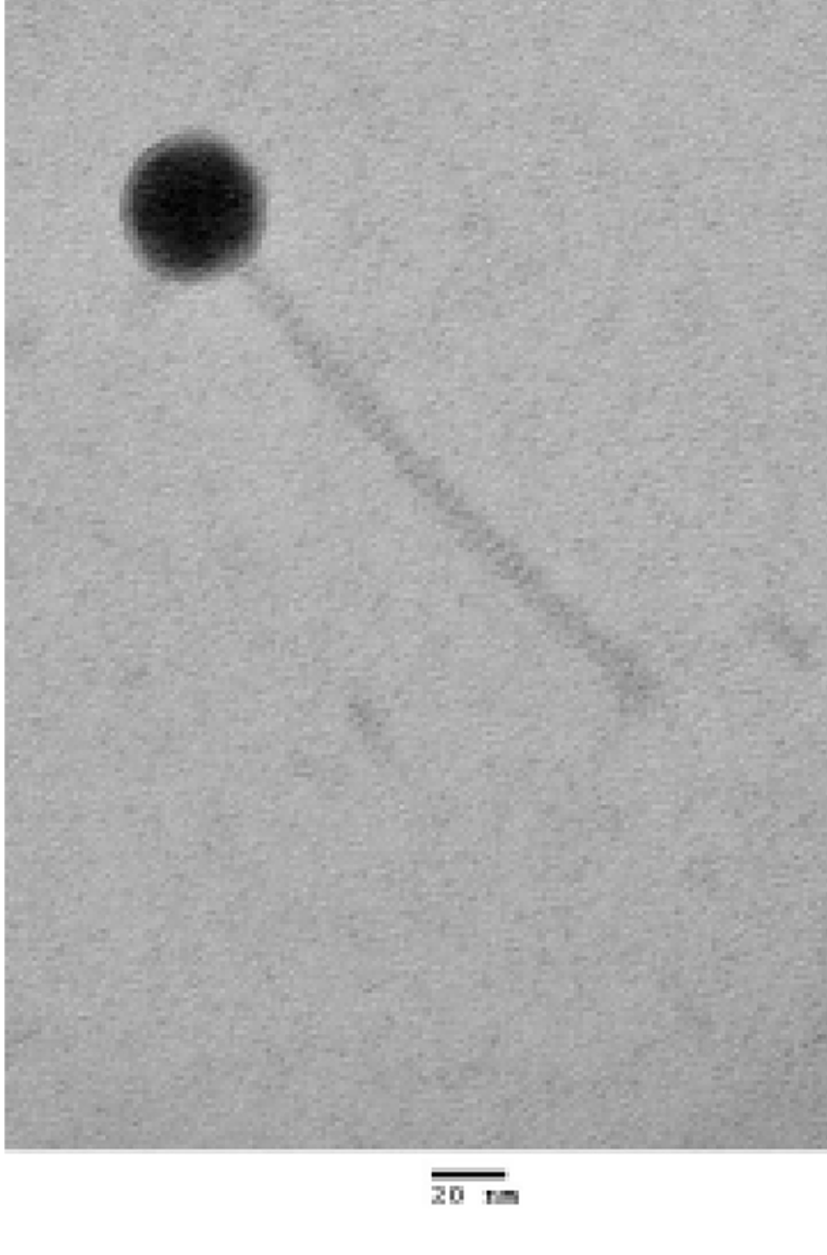
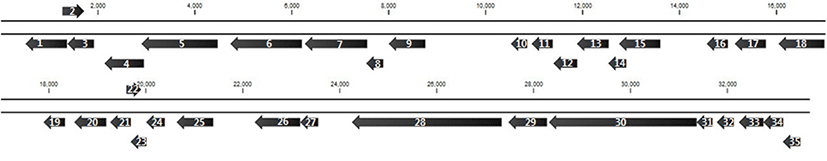
WCP30 phage was isolated from a fermented food and purified by plaque assay. To evaluate the morphology, WCP30 phage was observed by transmission electron microscopy (Fig. 1). According to the morphological analysis, WCP30 phage had a long non-contractile tail and an icosahedral head, thus it belonged to the family Siphoviridae in the order Caudovirales. Morphologically, phages can be tailed, polyhedral, filamentous, or pleomorphic; most of them contain a double-stranded DNA genome (Ackermann, 2003). The WCP30 phage genome was 33,697 bp in length, with 41.2% G+C content. The genome sequence were searched against GenBank by using BLASTN, and the results showed that WCP30 phage did not shared sequence of all phage for lactic acid bacteria. Bioinformatic analysis of the WCP30 phage genome revealed 35 putative ORFs. Of these, 20 ORFs had matches in GenBank with annotated functions. Comparison of the WCP30 phage sequence to sequences in GenBank showed that it was composed of the typical basic functional modules of phages, including replication, DNA packaging, stricture/morphogenesis, and lysis modules (Table 1 and Fig. 2).
In the structure/morphogenesis module, the predicted protein encoded by ORF1 was identified as the major capsid protein, as it showed similarity to the major head protein of Lactobacillus phage phiJL1. The ORF3 protein showed 44% similarity to a putative scaffolding protein, which is required for the correct assembly of coat protein and the incorporation of minor proteins. The ORF4 protein was identified as a head morphogenesis protein, as it showed 49% similarity to that of a phage infecting Leuconostocaceae bacterium R-53105. The ORF29 protein was identified as a tail protein, as it showed 45% similarity to a tail protein of W. paramesenteroides. The ORF33 protein was homologous (32% identity) to the major tail protein of Fructobacillus fructosus. The ORF30 protein, which was the longest ORF of WCP30 phage, showed sequence similarity to a tape measure protein (TMP) repeat in W. cibaria (68% identity). In a phage, the TMP determines the length of the tail by functioning as a template during tail assembly. Another structure/morphogenesis-related protein was encoded by ORF6. Within the DNA-packaging cluster, the proteins encoded by ORF7 and ORF8 were identified as a terminase and a small terminase subunit, respectively. The ORF5 protein showed sequence similarity (52% identity) with the phage portal protein from Leuconostocaceae bacterium R-53105. Together with the portal protein, the terminase, which is a protein complex composed of a large and a small subunit, drives DNA packaging (Moore and Prevelige, 2002). Within the lysis module, the ORF18 protein showed 43% similarity to the SGNH/GDSL hydrolase family protein of Fructobacillus ficulneus. ORF26 was predicted to encode a protein that is nearly identical to the N-acetylmuramoyl-L-alanine amidase from Leuconostoc citreum, and the ORF27 protein was identified as a putative holin, as it showed 23% similarity to a holin in a phage of Streptococcus mutans. Holins typically generate a lesion in the bacterial cytoplasmic membrane through which the endolysin passes (Gründling et al., 2001). In the replication module, ORF9 was predicted to encode a DNA (cytosine-5)-methyltransferase, as it showed 37% similarity to a DNA (cytosine-5)-methyltransferase of Leptotrichia goodfellowii F0264. ORF15 protein showed 26% similarity to the replicative DNA helicase of Bacteroides xylanisolvens, and ORF16 was predicted to encode a protein with 48% identity to the deoxyuridine 5'-triphosphate nucleotidohydrolase yncF of Marvinbryantia formatexigens. ORF 19 is predicted to encode a protein with 65% identity to a single-stranded DNA-binding protein of W. confusa, and ORF20 is predicted to encode a protein that is 51% similar to the SAK protein of the Lactococcus phage 936 group phage Phi129. The predicted ORF24 protein showed 69% identity to YopX of Fructobacillus sp. EFB-N1, and ORF 25 was predicted to encode a product that is homologous (69% identity) to the nicotinamide mononucleotide transporter of W. confusa. Nicotinamide mononucleotide transporter is integral membrane proteins that are involved in transport of nicotinamide mononucleotide (Fusco et al., 1990).
In conclusion, we analyzed the morphology and genome sequence of WCP30 phage, which was isolated from a fermented food. The genome sequence of this newly isolated WCP30 phage of W. cibaria was different from that of other phages that infect lactic acid bacteria. Further study is needed to determine the relationship between WCP30 phage and W. cibaria, especially when used as a starter for the fermentation of dairy products.