Introduction
Staphylococcus aureus is an important pathogen that causes food poisoning and nosocomial infections. Humans and animals are the main source of S. aureus. In humans, this bacterium can be normally present on the skin and in the oral and nasal mucosa. Thus, the colonized people are an important source for spreading staphylococci to others and to food. Also, colonization of animals with this bacterium can pose a potential risk for public health because it may result in contamination of milk and dairy products. S. aureus is an important cause of mastitis and this infectious disease is a very serious problem for dairy industry (Crago et al., 2012; Gundogan et al., 2005; Normanno et al., 2007; Peles et al., 2007).
S. aureus is an osmotolerant bacterium and can survive at low water activities for extended periods. However, S. aureus has a weakly competitive ability. Hence, this bacterium does not grow very well in food if there are low initial concentrations of staphylococci. Also, growth of this bacterium is easily suppressed by other microorganisms (especially lactic acid bacteria) in mixed cultures. However, S. aureus produces heat resistant enterotoxins, that cause food poisoning when growing in foods (Can and Çelik, 2012; Ertas et al., 2010; Jay et al., 2005; Pereira et al., 2009; Seo and Bohach, 2007; Tang et al., 2015).
Many researches have been conducted to investigate the presence of S. aureus in different foods. Milk, dairy products (cheese, ice cream), meats (beef, pork, turkey, chicken), eggs and egg products, and aquatic products are usually described as contaminated with S. aureus (Can and Çelik, 2012; Crago et al., 2012; Gücükoğlu et al., 2012; Gücükoğlu et al., 2013; Gündoğan et al., 2006; Hanson et al., 2011; Hu et al., 2013; Normanno et al., 2007; Pereira et al., 2009; Tang et al., 2015).
Because antibiotics are widely used in both clinical and agricultural fields, some foodborne pathogens display resistance to more than one antibiotic called ‘multidrug resistance’. Antibiotic resistance is usually genetically based, and S. aureus can gain resistance by mutation or conjugation (de Boer et al., 2009; McEntire and Montville, 2007; Pesavento et al., 2007; Smith and Jarvis, 1999; Tang et al., 2015).
Methicillin-resistant S. aureus (MRSA) has become a widely recognized cause of nosocomial infections world-wide. Although MRSA has a clinical importance due to its antibiotic resistance, the isolation of MRSA from foods has been reported. While the antimicrobial susceptibility profiles of clinical S. aureus isolates are well described, limited information about the antimicrobial susceptibility of foodborne strains of S. aureus is available in Turkey. The aims of this study were to evaluate the presence of S. aureus and MRSA in various food samples, and to determine the antimicrobial susceptibility and clonal relationships of the isolates.
Materials and Methods
A total of 160 samples were randomly collected between August 2014 and May 2015 in Hatay province, located in the southern Turkey. Samples including 50 raw milk (20 cows’, 15 goats’, and 15 ewes’ bulk milk), 50 traditional cheeses (25 Carra, 25 Surk), 30 chicken meat (wing and drumsticks with skin), and 30 beef minced meat were transported to the laboratory with ice packs for analysis.
Briefly, ten gram of each sample was added to 90 mL of 0.1% sterile peptone water (Oxoid, England), and homogenised using a stomacher (Bagmixer400, Interscience, France) for 2 min. Serial decimal dilutions were prepared from the mixture and 0.1 mL of appropriate dilutions were spread on Baird Parker agar (Oxoid) supplemented with Egg Yolk Tellurite Emulsion (Oxoid). Plates were incubated at 37°C for 24-48 h for enumaration of staphylococci. After the incubation period, up to five typical (grey/black colonies with precipitation zones) and/or atypical (black colonies with no zones) presumptive colonies were taken. These colonies were stored at −20°C in cryovials for coagulase activity, PCR analysis, and antimicrobial susceptibility test. A tube coagulase test was applied to these colonies using rabbit blood plasma with EDTA (Oxoid). Isolates indicated coagulation were considered CPS (Bennett and Lancette, 1998).
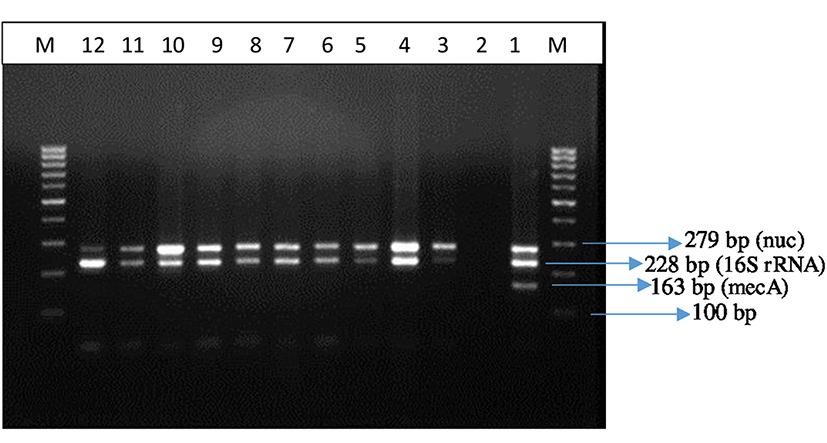
S. aureus ATCC 25923 was used as a positive control for targeting 16S rRNA and nuc gene. S. aureus ATCC 43300 was used as a positive control for mecA gene.
All coagulase positive isolates were identified by species-specific PCR. In this study, multiplex PCR assay was based on the detection of 16S rRNA (Staphylococcus genus specific), and nuc gene (S. aureus species specific). Methicillin resistance was also determined by testing for the presence of mecA gene. Specific primer pairs described by Brakstad et al. (1992), Mehrotra et al. (2000), Monday and Bohach (1999) were used. Primers (Ella Biotech GmbH, Germany) used in the study are shown in Table 1.
| Primer | Oligo nucleotide sequence (5’-3’) | Product size(bp) | Reference |
|---|---|---|---|
| 16S rRNA-F | GTA GGT GGC AAG CGT TAT CC | 228 | Monday and Bohach (1999) |
| 16S rRNA-R | CGC ACA TCA GCG TCA G | ||
| nuc-F | GCG ATT GAT GGT GAT ACG GTT | 279 | Brakstad et al. (1992) |
| nuc-R | AGC CAA GCC TTG ACG AAC TAA AGC | ||
| mecA-F | ACT GCT ATC CAC CCT CAA AC | 163 | Mehrotra et al. (2000) |
| mecA-R | CTG GTG AAG TTG TAA TCT GG |
DNA was extracted from the overnight Brain Heart Infusion broth (Oxoid) cultures by using a Bacterial DNA Extraction kit (Nucleic Acid Extraction Kit, Malaysia) according to the manufacturer’s protocol.
A multiplex PCR assay was performed in a reaction mixture of 25 μL final volume containing 1×PCR buffer (Thermoscientific, Lithuania), 1.5 mM MgCI2 (Thermoscientific), 200 μM of each dNTPs (EURx, Poland), 1 U Taq DNA polymerase (Thermoscientific), 0.12 μM of each primer, and 2 μL template DNA. Amplification conditions were performed as follows: initial denaturation at 94°C for 4 min and then 35 cycles of denaturation at 94°C for 30 s, annealing at 57.5°C for 30 s, and extension at 72°C for 40 s. Final extension cycle was performed at 70°C for 10 min (Keyvan and Özdemir, 2016; Mehrotra et al., 2000). Amplification products were detected by agarose gel (1.5%) electrophoresis carried out at 100 V for 50 min (CS-300V, England). The gels were stained with ethidium bromide and visualised under a UV transilluminator (UVP, USA).
The S. aureus isolates were tested for antimicrobial susceptibility by the disc agar diffusion method according to the guidelines of the Clinical and Laboratory Standards Institute (CLSI) (CLSI, 2013). The antibiotic discs were selected in line with the recommendation of CLSI and included penicillin (10 IU/disc), oxacillin (1 μg/disc), tetracycline (30 μg/disc), gentamicin (10 μg/disc), ampicillin (10 μg/disc), vancomycin (30 μg/disc), cefoxitin (30 μg/disc), chloramphenicol (30 μg/disc), erythromycin (15 μg/disc), ciprofloxacin (5 μg/disc), and rifampin (5 μg/disc). The isolates were classified as susceptible, intermediate resistant, and resistant.
Mulvey et al. (2001)’s protocol was used for the PFGE method. The dendogram analysis was made with Bionumerics program (Applied Maths, Inc., Belgium, 6.01 version). Tenover et al. (1995)’s criteria were used for detecting the clonal relation among the S. aureus strains.
Results and Discussion
A total of 160 food samples were examined for the presence of S. aureus and MRSA using multiplex PCR analysis. Twenty (12.5%) of the samples were found to be contaminated with S. aureus. A total of 40 isolates from the 20 positive samples were confirmed to be S. aureus by multiplex PCR based on 16S rRNA and nuc gene. Among them 25 were from raw milk, 4 were from Carra cheese, and 11 were from chicken meat samples. The mecA gene was not detected in any of the S. aureus strains.
When we examined the studies conducted in order to determine the presence of S. aureus in various foods in Turkey, the contamination levels with S. aureus of the analysed food samples have been observed to be 53.3% by Gundogan et al. (2005), 61.1% by Gündoğan et al. (2006), 57.3% by Ertas et al. (2010), and 52.4% by Gücükoglu et al. (2012), which are quite higher than our results. Nevertheless, Can and Çelik (2012) detected S. aureus in only 6% of the cheese samples they had analysed, and this rate is lower than the rate we had found in our study.
Moreover, in this context, when the studies conducted in other countries are considered, it is observed that the contamination levels are different from each other, and S. aureus is usually isolated from foods which are rather rich in proteins. Presence of S. aureus was reported to be 12.8% in meat and dairy samples by Normanno et al. (2007), 15.6% in various food samples by Hu et al. (2013), and 16.4% in raw meat samples by Hanson et al. (2011), and these levels are close to the contamination level (12.5%) we had found in the present study. Unlike the aforementioned findings, the contamination levels with S. aureus have been found to be 31.9% in tank milk samples by Muehlherr et al. (2003), 23.8% in raw meat by Pesavento et al. (2007), and 85.1% in different foods in China by Tang et al. (2015). However, S. aureus was isolated from the 10.5% of the food samples held responsible for the foodborne infections by Crago et al. (2012) in Canada. The differences with the results of contamination levels with S. aureus may result from the fact that some of the foods tested in our study are traditional and have different production techniques. In addition to this, it could be also linked to the differences in personal hygiene, milking hygiene and slaughter hygiene through the food chain.
The antibiotic susceptibility profile of the isolates to 11 different antibiotics was tested; all isolates were susceptible to gentamicin, oxacillin, and vancomycin. The highest resistance rate was detected in penicillin (95%) and ampicillin (92.5%), followed by tetracycline (30%), erythromycin (20%), ciprofloxacin (12.5%). The lowest rate of resistance was found to be against chloramphenicol (2.5%), and cefoxitin (2.5%) (Table 2).
In this study, 39 out of 40 (97.5%) isolates were found to be resistant to one or more antibiotics. Only one isolate from chicken meat was susceptible to all the tested antibiotics. All isolates from raw milk and Carra cheese were resistant to penicillin and ampicillin.
Among the isolates collected from chicken meat, 9.0% (1/11) of them were resistant to chloramphenicol, 72.7% (8/11) were resistant to ampicillin, 45.4% (5/11) were resistant to tetracycline, 9.0% (1/11) were resistant to cefoxitin, 36.3% (4/11) were resistant to erythromycin, 81.8% (9/11) were resistant to penicillin, and 27.2% (3/11) were resistant to ciprofloxacin. In addition, two isolates (18.1%) were found to be intermediately resistant to ciprofloxacin. In this study, isolates from chicken meat were more resistant to antibiotics than the isolates collected from raw milk and Carra cheese. This difference may be due to the widely use of antibiotics for therapeutic and prophylactic purposes at poultry farms in Turkey.
When the results of antibiotic sensitivity test are examined, higher resistance rates were found against beta-lactam antibiotics, penicillin and ampicillin among the isolates from raw milk and Carra cheese. Since beta-lactams have an important place in the treatment of mastitis in Turkey, this could be explained by the extensive and uncontrolled use of these group antibiotics in agriculture. However, no isolate was found to be resistant to methicillin in this study. Apart from the penicillin resistance, Gundogan et al. (2005), and Gündoğan et al. (2006) found that bacitracin and methicillin resistance was prevalent among the isolates they had obtained. The sensitivity rates of vancomycin, oxacillin, gentamicin, and rifampin we had detected in the present study were in compliance with the rates reported in some other studies (Can and Çelik, 2012; Gundogan et al., 2005; Gündoğan et al., 2006; Hanson et al., 2011; Hu et al., 2013; Normanno et al., 2007; Pesavento et al., 2007).
In contrast, 38% of the isolates were found to be resistant to oxacillin in a study conducted in Portugal (Pereira et al., 2009). Peles et al. (2007) found the S. aureus strains isolated from the milk of mastitic cows, and the bulk tank milk to be sensitive to methicillin, cefoxitin, tetracycline and erythromycin, while penicillin resistance was found to be prevalent among the isolates in a similar manner. The fact that the isolates were found to be highly resistant (83.3%) to one or more of the antibiotics tested in a study carried out by Can and Çelik (2012) shows similarity with the findings of our study. Unlike our study, some researchers found MRSA in raw meat, chicken meat, raw milk, dairy and soy products (Can and Çelik, 2012; de Boer et al., 2009; Gundogan et al., 2005; Gündoğan et al., 2006; Gücükoğlu et al., 2013; Hanson et al., 2011; Pereira et al., 2009; Tang et al., 2015).
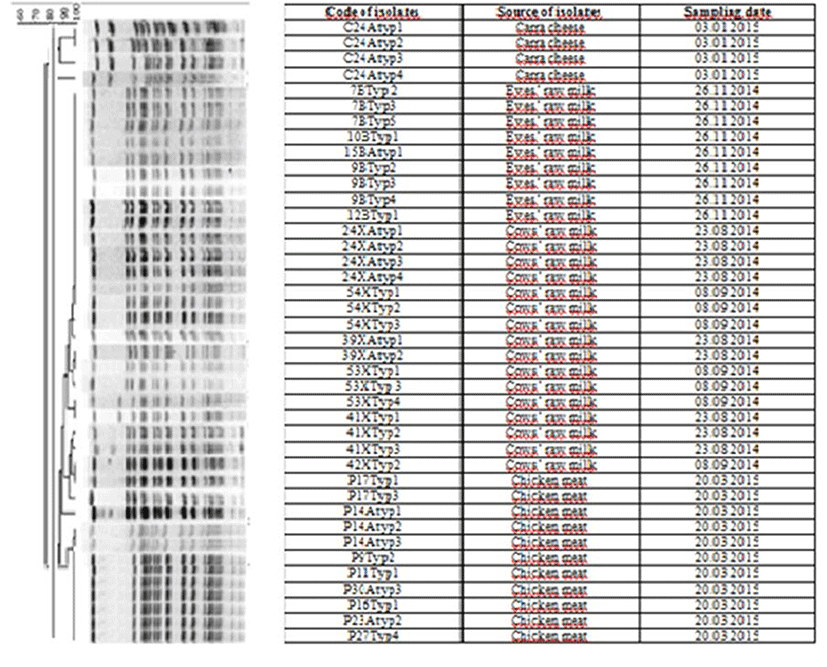
In this study, dendogram of PFGE patterns of S. aureus strains was also performed. SmaI restriction enzyme was used in PFGE method. Nine major patterns were determined in 40 strains. In 3 of the patterns there was a strain closely related to the main cluster. In 6 of the patterns, the number of strains changed between 2-16 and thirty-six strains (90%) had identical PFGE profiles. Two major groups were detected when the similarity coefficient was evaluated as 85% and above. The first group was consisted of 4 strains with a similarity rate of 88%, and the second group was consisted of 36 strains with a similarity rate of 90% (Fig. 2).
Conclusion
These results point out that S. aureus strains resistant to antibiotics may be not only nosocomial borne, but also present in food. Although our study is a limited research about the occurence of S. aureus in different foods, these analysed foods are usually consumed by local people in this area. As a result, we think the fact that resistance rates against tested antibiotics among the isolates were found to be high, and the multidrug-resistant S. aureus strains were also found is an important problem that should be overemphasised in terms of public health and food safety.