Introduction
Muscle protein oxidation occurs in a number of diseases of living tissues (Coirault et al., 2007; Oh-Ishi et al., 2003) as well as in muscle foods during meat-processing and storage (Lund et al., 2011; Soladoye et al., 2015). Reactive species, such as hydroxyl, superoxide, peroxyl, nitric oxide, hydrogen peroxide, and peroxynitrite, as well as transition ions, are major factors in protein oxidation (Lund et al., 2011; Shacter, 2000; Stadtman, 1993). These factors can lead to structural and biochemical changes of muscle proteins with modifications of the amino acid side-chains and polypeptide backbones of the muscle proteins (Lund et al., 2011). Consequently, protein oxidation affects the deterioration of quality in muscle foods and the consumer’s health.
To delay or prevent oxidation in muscle foods requires an understanding of the various mechanisms of protein oxidation, such as carbonylation, metal ion-catalyzed oxidation, cross-linking, hydroxylation, sulfoxidation, and nitrosylation. A number of studies have been conducted to define these mechanisms (Armenteros et al., 2009; Decker et al., 1993; Garrison, 1987; Levine, 1984; Stadtman and Berlett, 1988; Stadtman and Levine, 2003). Various methods for assessing protein oxidation have also been introduced. The most commonly used method for measuring total carbonyls derived from proteins is the dinitrophenylhydrazine (DNPH) method (Oliver et al., 1987). Thiol oxidation, which is disulfide bond-formation in thiol-containing amino acid (cysteine) residues, can be determined using dithiobisnitropyridine (DTNP) (Winterbourn, 1990). Daneshvar et al. (1997) introduced α-aminoadipic semialdehyde (AAS) and γ-glutamic semialdehyde (GGS) as biomarkers of oxidative lysine and arginine (or proline). Recently, proteomic and spectrometric technologies have been applied in studies on specific amino acid oxidation. Bernevic et al. (2011) and Ghesquiere et al. (2011) assessed oxidized methionine, whereas Jongberg et al. (2011) and Estévez et al. (2009) applied LC-MS to the detection of potential thiol-quinone adducts and AAS and GGS, respectively. Although the proteomic approach allows the study of oxidation of specific amino acids and proteins, few studies have been conducted on muscle foods: arginine and proline (Stadman and Levine, 2003), cysteine (Lund et al., 2011), glutamic acid and tyrosine (Xiong, 2000).
In the present study, the myosin protein of porcine skeletal muscle was used for studying methionine oxidation. Myosin is the most abundant protein distributed in skeletal muscles and plays an important role in muscle contraction and metabolism due to the existence of a motor domain and an actin-binding site (Rayment et al., 1993; Tokunaga et al., 1987). Thus, myosin protein has been widely used in studies on muscle protein proteolysis, denaturation, and oxidation, as well as on its relationship with meat quality and carcass trait in livestock (Coirault et al., 2007; Hanan and Shaklai, 1995; Kim et al, 2013; Lund et al., 2008). In our previous report, we established that LC-MS/MS was a useful approach to the analysis of myosin isoforms from bovine skeletal muscle. LC-MS/MS can also be used to assess methionine oxidation by identification of mass modification (Bernevic et al., 2011; Kim et al., 2015). Therefore, the purpose of this study was to investigate the oxidized methionine of porcine myosin using proteomic technology.
Materials and Methods
Three castrated pigs ((Landrace × Yorkshire) × Duroc) were slaughtered at the body weight of 108±2.5kg (182 d of age) by electrical stunning in a commercial slaughter house. Immediately after slaughter, three muscles (longissimus thoracis, LT; psoas major, PM; semimembranosus, SM) were taken from each pig within 45 min postmortem. Muscle samples were soaked in lysis buffer consisting of 7 M urea, 2 M thiourea, 1% (w/v) DTT, 4% (w/v) CHAPS, and 0.03 M Tris, and subsequently transferred to a laboratory. Samples were homogenized using a polytron homogenizer (IKA Labortechnik T25-B, Malaysia) at 3,000 rpm for 25 s. The homogenates were centrifuged at 15,000 g at 4℃ for 60 min. Eluted fractions were precipitated by trichloroacetic acid/acetone precipitation and the pellets were dissolved in a lysis buffer. The protein concentration was adjusted to 1.0 mg/mL by using the Bradford (1976) method with bovine serum albumin as a standard. In-solution digestion was conducted using trypsin (Promega, UK). The protein sample was digested in a trypsin solution (2.5 ng/μL) in 50 mM NH4HCO3 buffer (pH 7.8) at 37℃ for 12 h.
Protein digests were analyzed using a nano-LC and LTQ mass spectrometer (Applied Biosystems, USA). Samples were injected into a 75 μm × 150 mm column (Proxeon, Denmark) and the capillary column was slurry packed inhouse with 5 μm and 100 Å pore-size Magic C18 stationary phase (Michrom Bioresources, USA). For the LC separation, mobile phase A was 0.1% formic acid in deionized water and mobile phase B was 0.1% formic acid in acetonitrile. Peptides were separated using 75 min gradients (5-35% B for 50 min, 40-60% B for 20 min and 60-80% B for 5 min) and the flow rate was maintained at 300 μL/min after splitting. Mass spectra were acquired from m/z 400-1800 followed by MS/MS scans, and each MS/MS scan acquired was an average of one microscan on the LTQ. The temperature of the ion transfer tube was controlled at 200℃ and the spray was 1.5-2.0 kV. The normalized collision energy was set at 35% for the MS/MS. Peptide sequences were identified via the NCBInr (Sus scrofa) database, searching for a peak list using the MASCOT algorithm (Matrix Science, UK). The database search criteria were as follows: fixed modification, no; variable modification, oxidized at methionine residues; maximum allowed missed cleavage, 1; peptide MS tolerance, 100 ppm; fragment MS tolerance, 0.1; peptides resulting from trypsin digest were considered. The ion score was -10×log(P), where P is the probability that the observed match is a random event. Individual ion scores of >39 indicated identity or extensive homology (p<0.05). All of the identified peptides with ion scores 39 were deleted from the list.
Results and Discussion
Four myosin isoforms (myosin-1, -2, -4, and -7) were identified in porcine LT, PM, and SM muscles by LC-MS/MS analysis (Table 1). The number of queries matched to the amino acid sequences of myosin isoforms ranged from 72 to 237 and the mean value was 157.9. The average of sequence coverage was 38.0%. It is necessary to check the unique peptides identified from each myosin isoform because of high homology among these isoforms. In the present study, 7.3-13.3 of the unique peptide corresponding to each isoform was detected from three porcine muscles. Although it is impossible to quantitatively analyze the myosin isoforms from the results of this study, we can confirm which isoforms were distributed in the muscle samples. As presented in Table 2, each unique peptide can be conclusive proof of identification of each myosin isoform. Moreover, these peptides were detected in all three porcine muscles (LT, PM, and SM). Commonly expressed peptides in two or more myosin isoforms supported the distribution of specific myosin isoforms (Table 3). All queries except for DLEEATLQHEATAAALR were peptides composed of myosin-1. These queries were also observed as part of myosin-2 or -4, whereas only four queries were peptides composed of myosin-7.
a)Accession numbers were taken from the NCBI database.
b)Theoretical molecular weight.
c)Peptide size (amino acids) recorded in NCBI database.
d)LT, longissimus thoracis; PM, psoas major; SM, semimembranosus.
a)Individual ion score > 39 indicates identity (p<0.05).
a)Individual ion score > 39 indicates identity (p<0.05).
In our previous report, three myosin isoforms (myosin-1, -2, and -7) were identified from bovine longissimus muscle by analysis of LC-MS/MS (Kim, 2014). However, additional analyses were needed because it was not confirmed whether myosin-4 existed in this muscle. Although no unique queries of myosin-4 were detected, some queries were identified as peptides expressed in common in two or more isoforms including myosin-4. A number of previous reports that studied myosin isoforms using various technologies, such as electrophoresis, histochemistry, immunohistochemistry, immunoblotting, and RT-PCR in various breeds of pigs, supported the clain that there are four myosin isoforms in porcine skeletal muscle (Abreu et al., 2006; Chang et al., 2003; Kim et al., 2014; Lefaucheur et al., 2002). It has been observed that the composition of myosin-7 (myosin heavy chain I/slow) in porcine skeletal muscle was the lowest among the myosin isoforms at the mRNA and protein levels (Choi et al., 2007; Park et al., 2009). In this study, myosin-7 had lower values in matched queries and in sequence coverage compared to the others.
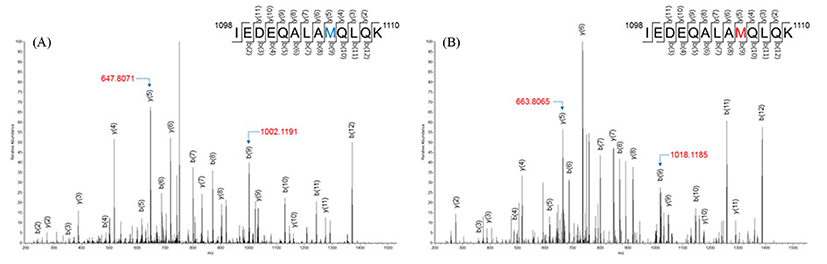
Five methionine-oxidized peptides were detected among 836 trypsin-digested peptides, as shown in Table 4. SM muscle had all of the methionine-oxidized peptides, whereas PM and LT had one and two, respectively, and these oxidized peptides had one or two oxidized methionines. Trypsin-digested peptides generally had a methionine, and their oxidation could be identified by mass modification. Peptide IEDEQALAMQLQK, which corresponded to residues 1098-1110 of myosin-1 (or residues 1096-1108 of myosin-4), had an oxidized methionine. The MS/MS spectra of this peptide showed 16 amu of mass modification in residue 106 (Fig. 1). The mass of y(5) corresponding to 1106MQLQK1110 was m/z 647.8071 (Fig. 1A), whereas the spectrum of oxidized peptide corresponding to 1106MQLQK1110 was observed at 16.0 amushift (m/z 663.8065) (Fig. 1B). The mass of b(9) also showed the same mass shift (m/z 1002.1191 to m/z 1018.1185) in 1098IEDEQALAM1106 (Fig. 1).
a)LT, longissimus thoracis; PM, psoas major; SM, semimembranosus.
b)Individual ion score > 39 indicates identity (p<0.05).
c)Myosin isoform (residues).
M on the bar indicates oxidized methionine.
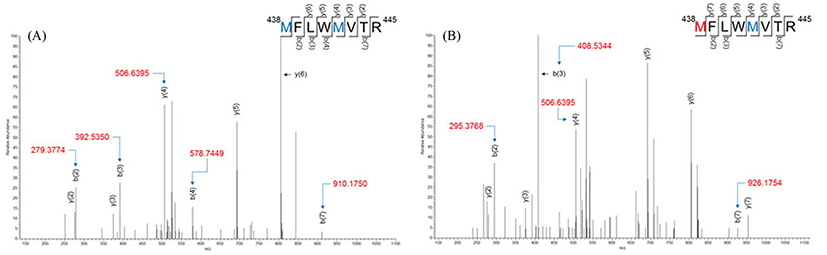
MFLWMVTR, which corresponded to residues 438-445, had two methionines (residues 438 and 442). As presented in Fig. 2, the methionine corresponding to residue 438 was identified as methionine sulfoxide (the addition of one oxygen), whereas another methionine (residue 442) did not oxidize. The masses of b(2) and b(3) from non-modified peptides (Fig. 2A) were m/z 279.3774 and m/z 392.5350, respectively, whereas those from modified peptides (Fig. 2B) were m/z 295.3768 and m/z 408.5344, respectively. Thus, 16 amu of mass shift was confirmed by the mass of b(2) and b(3). The b(7) corresponding to 438MFLWMVT445 indicates that methionine on residue 442 did not oxidize due to the mass shift, was 16.0amu (m/z 910.1750 to m/z 926.1754). Moreover, there was no mass shift of y(4) between non-oxidized peptide (m/z 506.6395) and oxidized peptide (m/z 506.6395).
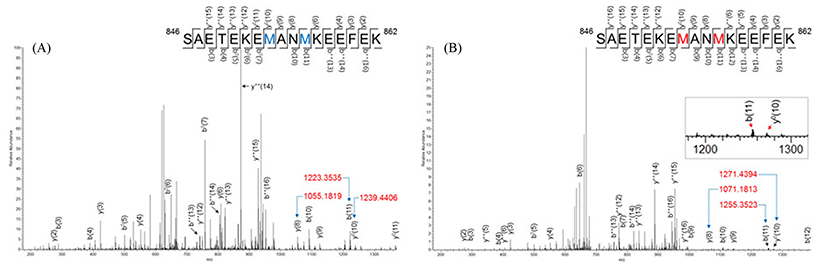
Two methionine oxidations were found in SAETEKEMANMKEEFEK that corresponded to residues 846-862 of myosin-1 (or residues 844-860 of myosin-4) (Fig. 3). The mass difference of b(11) between non-oxidized peptide (m/z 1223.3535) and oxidized peptide (m/z 1255.3523) was 32.0amu. The mass shift of b(11) indicated that two oxygens were added to this peptide, and three cases are predictable as follows: the methionine on residue 853 oxidized to the sulfone form, another methionine (on residue 856) formed a methionine sulfone, and both of the methionines oxidized as sulfoxide forms. The y(8) corresponding to 855NMKEEFEK862 showed 16.0 amu of mass shift, while 32.0 amu of mass shift was found for the y0(10) corresponding to 853MANMKEEFEK862.
It is generally accepted that protein-bound methionine easily forms methionine sulfoxide by reacting with peroxides, and methionine sulfone may also often be formed (Vogt, 1995). In the present study, only methionine sulfoxide was detected. Very little oxidized methionine was identified in porcine skeletal muscle, and this may be due to the early collection of muscle samples (within 45 min postmortem). In the muscle-food system, myofibril proteins are affected by various oxidants during processing and storage (Martinaud et al., 1997). In addition, the oxidation of methionine and other amino acids can be affected by neighboring amino acids (Ghesquiere et al., 2011). Methionines surrounded by acidic amino acids, such as aspartic acid (D) and glutamic acid (E), or those surrounded by alanine (A), threonine (T), and serine (S), are susceptible for oxidation. In the present study, there were A or T nearby oxidized methionines on residues 853, 1071 and 106 of myosin-1.
It is known that the general oxidative changes in proteins are protein cross-linking, modifications of amino acid side chains and protein fragmentation caused by the formation of disulfide bonds, hydroperoxide, carbonyl, sulfoxide, and sulfone groups, as reported by Lund et al. (2011). These oxidants lead not only to reducing the water-holding capacity and the tenderness of meat, but also the deterioration of protein functionality, such as in hydrophobicity, conformation, and solubility (Lund et al., 2007; Rowe et al., 2004). The effect of methionine oxidation on cellular signaling was the main focus in previous reports (Erickson et al., 2008; Hardine et al., 2009). However, little attention was paid to the impact of methionine oxidation in food systems.
All of the oxidized peptides identified in porcine skeletal muscles were commonly-expressed peptides in two or three myosin isoforms, while there was no unique peptide expressed in one myosin isoform among the oxidized peptides. In particular, no peptides of myosin-7 were detected as oxidized peptides, regardless of whether they were unique or common peptides. This may be due to the different compositions of the myosin isoforms of the muscle sample. The content of myosin-7 is the lowest among the myosin isoforms in porcine skeletal muscles (Choi et al., 2007; Park et al., 2009). In this study, we did not quantified each myosin isoform, but it was estimated that the content of myosin-7 was lower than the others because of the lowest number of matched queries, as shown in Table 1. Protein oxidation can occur by chemicals and equipment during analytical steps (Chen and Cook, 2007; Liu et al., 2013). Nevertheless, LC-MS/MS is better suited for studying protein oxidation than two-dimensional gel electrophoresis (Ghesquière and Gevaert, 2014). Liu et al. (2013) presented the accurate determining method for methionine oxidation by stable isotope labeling and LC-MS analysis. In the present study, although overestimation of oxidized methionine level was not excluded, oxidized methionine can be detected from porcine skeletal muscles by LC-MS/MS analysis.
Conclusions
Five peptides with one or two oxidized methionines were detected among 836 queries of myosin isoforms from porcine skeletal muscles, and these were identified by LC-MS/MS. All oxidized methionines were identified in the methionine sulfoxide form by an oxygen modification (16 amu-shift), while methionine sulfone was not observed in the present study. The LC-MS/MS approach can also allow the analysis of methionine oxidation on specific residues and specific myosin isoforms in porcine skeletal muscle.













