Introduction
Synthetic nitrite is an essential additive in processed meat products for meat color development, Clostridium botulinum decontamination, and the enhancement of curing, flavor, and antioxidant effects. In the late 1960s and early 1970s, when processed meat products including bacon and ham are cooked at high temperature, synthetic nitrite was reported to react with amines to form nitrosamines, some of which are carcinogenic, as reported in animal studies (Gray et al., 1981). Moreover, nitrite overuse may oxidize hemoglobin, causing various side-effects including met-hemoglobinemia (Glandwin et al., 2004). Therefore, the advantages and disadvantages of synthetic nitrites have remained controversial since the 1970s until today, and currently numerous countries worldwide have imposed restraints on the use of synthetic nitrite (Honikel, 2008).
Concurrent with the health-oriented consumption patterns of modern consumers and the negative perception of synthetic additives, numerous studies have attempted to identify an alternative to synthetic nitrite (Sebranek and Bacus, 2007; Viuda-Martos et al., 2009). In the 1990s, companies began developing new methods for curing meat with celery or other natural nitrate/nitrite sources. Accordingly, two methods were proposed: one based on direct substitution of each nitrite function in meat products with an alternative material and the other based on indirect substitution where nitrite-rich vegetables are used as the source and nitrate reductase-producing microorganisms are cultured to mediate the conversion from nitrate to nitrite (Hammes, 2012).
The method based on indirect substitution of synthetic nitrite is currently being used in the meat industry here and abroad (Alahakoon et al., 2015). Processed meat products, for which the conversion of high nitrate levels in vegetable powder or extract (approximately 30,000 ppm) to nitrite via microbial fermentation, have been developed and commercialized, where the relatively expensive vegetable powder and the fermentation microorganism needed for nitrate reduction are mostly imported from multinational corporations (Sindelar, 2006). Furthermore, vegetables used in this method, including celery and beet, reportedly impart a strong and distinct flavor to meat products and reduce palatability among Korean consumers with limited exposure to foreign flavors. While synthetic nitrite is indeed essential for preventing food poisoning caused by Clostridium botulinum and for color development in meat products (Kim et al., 2016), consumers repeatedly avoid them. Naturally occurring nitrate is anticipated to replace nitrite with domestically grown vegetables being standardized and added to meat products in lieu of nitrite additives (Riel et al., 2017). Thus, a nitrite substitution method customized in accordance with Korean standards should be developed, and a method of replacing expensive imported materials should be developed. Furthermore, selection of the fermentation microorganism with nitrate reductase activity is a prerequisite for converting nitrate in enriched vegetable powder or extract to nitrite.
This study applied kimchi-derived microorganisms used for a culture starter and an alternative to synthetic nitrite in meat products, as they can grow under conditions of low temperature and certain salt concentrations and in the presence of materials containing either nitrate or nitrite.
Materials and Methods
Nitrate-rich vegetable-based kimchi: cabbage kimchi, spinach kimchi, leaf mustard kimchi, turnip kimchi, young radish kimchi, and cubed radish kimchi, were transferred into a sterile stomacher bag with 90 mL of a sterile 0.85% NaCl solution and then mixed for 5 min in a stomacher, respectively. After 10-fold serial dilutions of 1 mL of the suspension, the diluents were spread onto De Man, Rogosa, and Sharpe (MRS) agar supplemented with nitrite (200 ppm) and cultured at 30°C for 48 h.
Nitrite-resistant isolates from various types of kimchi and kimchi lactic acid bacteria obtained from Microorganism and Gene Bank (MGB) were cultured in an MRS broth supplemented with 200 ppm nitrate (NaNO3) at 30°C for 48 h. After centrifugation (8,000×g for 15 min at 4°C), nitrite levels in the culture supernatant were determined using a nitrite high-range portable photometer (Hanna Ins., Woonsocket, RI, USA) for initial screening of the kimchi-fermenting microorganisms producing high levels of nitrite. These bacteria were then cultured in a BBL-indole nitrate medium at 25°C for 36 h, and nitric oxide levels in the culture supernatant were determined using a Griess reagent kit (Thermo Scientific, Waltham, MA, USA) in accordance with the manufacturer’s instructions, and the absorbance was measured at 548 nm.
The cellular phenotype of the strains was examined using the method of published paper (Logan and Berkeley, 1984). Vegetative cells were observed using a phase-contrast microscope (Nikon, Tokyo, Japan). Gram staining was performed using a Gram staining kit (BD Difco, NJ, USA). Growth at pH 2.0–13.0 (at intervals of 1.0 pH unit) was determined in trypticase soy broth (Difco) adjusted with citrate/phosphate or Tris-HCl buffers. Growth at different temperatures (10°C, 20°C, 30°C, 40°C, 50°C, and 60°C) and with 0%–15% (w/v) NaCl (at intervals of 1% NaCl; 30°C) was assessed on TSA for 4 d. Growth under anaerobic conditions was assessed on TSA at 30°C, using a GasPak jar (Merck Millipore, Burlington, MA, USA) for 4 d. Biochemical assays for phenotype characterization were performed at 30°C, using API 50 CH strips with API 50 CHL medium (bioMérieux, Lyon, France), in accordance with the manufacturer’s instructions.
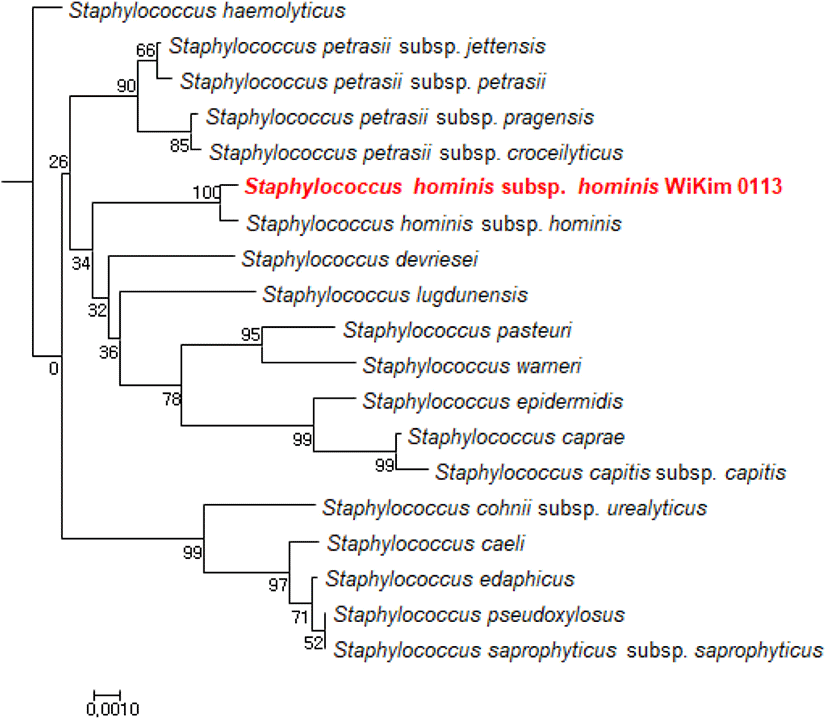
Polymerase chain reaction (PCR) was performed (Minicycler, MJ Research Inc., Waltham, MA, USA) to amplify a partial 16S rRNA fragment from the isolated strain using universal primers (27F: 5'-AGAGTTTGATCATGGCTCAG-3' and 1492R: 5'-GGATACCTTGTTACGACTT-3'). The cycling conditions were as follows: initial denaturation at 95°C for 5 min; followed by 35 cycles of denaturation at 94°C for 45 s, annealing at 52°C for 45 s, extension at 72°C for 1 min; final extension at 72°C for 5 min. The amplified PCR product was ligated into a T vector (Invitrogen, Carlsbad, CA, USA). 16S rRNA sequencing was performed using an ABI 377 Genetic Analyzer (Applied Biosystems, Foster, CA, USA). The 16S rRNA gene sequences from the isolates were aligned with GenBank reference sequences (http://www.ncbi.nlm.nih.gov) using the Basic Local Alignment Search Tool (BLAST) to identify the taxonomic position of bacterial strains. Multiple sequence alignments were performed using CLUSTAL_W (Thompson et al., 1997), and alignment positions with gaps and unidentified bases were excluded using BioEdit. A phylogenetic tree was constructed using the neighborhood-joining method and bootstrap percentages based on 1,000 replications (Saitou and Nei, 1987). MEGA 4.0 was used to assess the phylogenetic tree.
To assess the hemolytic activity, the selected strain was streaked onto blood agar media, containing 5% (v/v) of sheep blood and incubated at 30°C for 24 h. The extent of hemolysis was examined through the formation of a zone of clearance around the colonies (β-hemolysis, clear zones; γ-hemolysis, no zone). Proteolytic activity was assessed using skim milk agar (2% skim milk and 1.5% agar). Five microliters of the culture of the selected strain was spotted on skim milk agar and incubated at 30°C for 48 h. Proteolytic activity was examined by observing the formation of the zone of clearance around the colony.
Enzymatic characterization of selected strain was carried out using a semi-quantitative API ZYM kit (BioMérieux, Marcy-I’Etoile, France). The experiment was performed in accordance with the manufacturer’s instructions. Cultures of strains were centrifuged (8,000×g for 15 min at 4°C), and the pellets (106 CFU/mL) were placed in individual cupules through reattachment to sterilized 0.85% NaCl solution. Briefly, the microcupules of the API-ZYM strip were inoculated with 24-h-old broth culture of selected strain and incubated at 30°C for 4 h. After incubation, ZYM A and ZYM B reagents were consecutively supplemented to each cupule. Finally, API-ZYM strip was exposed to light. Progression of substrate hydrolysis (nmol of product) was examined on the basis of the intensity of color change. Grades 0 and 1 were considered negative and grades 2, 3, 4, and 5 were considered moderately-to-strongly positive.
Antibiotic susceptibility of indicator bacteria was carried out using trypticase soy agar by agar disc diffusion method. Indicator bacteria was aseptically streaked on TSA using sterile swab. The following antibiotics discs (BD BBL, Franklin Lakes, NJ, USA) were then placed on the surface of the solidified agar and allowed to diffuse into the agar for 10–15 minutes before incubating at 30°C for 24 h : Ampicillin (10 μg), Cefotetan (30 μg), Chloramphenicol (30 μg), Ciprofloxacin (5 μg), Clindamycin (2 μg), Gentamicin (10 μg), Doxycycline (30 μg), Erythromycin (15 μg), Kanamycin (30 μg), Penicillin G (6 μg), Streptomycin (10 μg), Tetracycline (30 μg), Trimethoprim-sulfamethoxazole (25 μg), Vancomycin (30 μg).
The agglutination test was performed by Pastorex Staph-Plus (Bio-Rad, Marnes la Coquette, France) according to manufacturer's instructions. A few colonies of Staphylococcus spp. were placed into a marked black circle on the Pastorex Staph-plus reaction card. A drop of the latex reagent was added inside circle of the card and colonies inside were thoroughly mixed with a wooden applicator stick. The card was rotated and examined for 20 seconds. A positive reaction was defined as clumping of the latex particles with substantial clearing of the milky background. Staphylococcus aureus ATCC25923 was used to the interpretation of a positive agglutination test. The indicator strain was obtained from the American Type Culture Collection (ATCC), and then cultured in tryptic soy medium (Difco, Franklin Lakes, NJ, USA) for 24 h at 37°C. Agglutination was scored as positive (+), equivocal (+), or negative (–). Latex particles sensitized by bovine albumin solution as negative controls.
For detection of enterotoxin genes in Staphylococci cultures, colonies were harvested from tryptic soy agar. To directly extract the microbial DNA, 3-mL aliquots of the suspension of colonies were centrifuged at 8,000×g for 15 min. The pellets were subjected to automated QIAcube extraction using QIAamp DNA mini kit (Qiagen, Hilden, Germany) in accordance with the manufacturer’s instructions. The concentration and purity of the extracted DNA were determined using a Nanodrop 2000 (Thermo Scientific). The purified DNA samples were stored at −20°C. Primers used for the detection of SEB, SEC and TSST-1 were as described previously (McLauchlin et al., 2000). The sequences of all primers together with their respective amplified fragments are summarized in Table 1. According to the manufacturer recommended protocol, PCR was carried out using commercially available PCR premix (AccuPower PCR PreMix, Bioneer, Korea), which contained 15 ng DNA template and 2 μL of primer set (10 pmol). DNA amplification was performed in a thermal cycler (Eppendorf, Hamburg, Germany) with initial denaturation at 94°C for 5 min followed by 35 cycles of denaturation at 94°C for 1 min, primer annealing 55°C for 1 min and extension at 72°C for 1 min, followed by a final extension at 72°C for 7 min. The amplified PCR products were electrophoresed on 1% agarose gels with 1× TAE buffer (20 mM Tris, 10 mM sodium acetate, 0.5 mM Na2EDTA, pH 8.0), stained with loadingSTAR (DyneBio, Seoul, Korea), and then visualized by ChemiDoc UV transilluminator (Biorad, CA, USA). Staphylococcus aureus ATCC25923 reference strain was used as an enterotoxigenic positive control.
Clostridium perfringens KCCM13124 was used to determine the antibacterial activity of the selected strain. This indicator strain was obtained from the Korean Culture Center of Microorganisms (KCCM) and then cultured in a reinforced clostridial medium at 37°C for 24 h. Antibacterial activity of culture supernatants against Clostridium perfringens (causing food poisoning) was assessed using the agar well diffusion assay, using a previously described method (Schoster et al., 2013). Selected strains were incubated in MRS medium supplemented with nitrate (200 ppm) at 30°C for 48 h. Culture supernatants were obtained through centrifugation (8,000×g for 20 min at 4°C) and used as an antagonistic substance. Wells (8 mm) impregnated with 120 μL of culture supernatant were placed on reinforced clostridial agar plates seeded with C. perfringens (107 CFU/mL) and subsequently incubated under anaerobic conditions at 30°C for 48 h. Fermentation and non-fermentation samples were subsequently aliquoted to these wells. The diameter of each zone of clearance was measured in millimeters to assess the antagonistic effect of the selected nitrite-producing strain.
Minimal growth medium used herein for nitrate reduction was used under similar conditions as meat products. Peptone-beef (PB) medium was prepared through nutrient supplementation including 0.1% glucose, 1% beef extract, 1% peptone, 0.01% iron chloride, 0.01% iron sulfate, 0.01% molybdenum oxide, and 0.02% sodium nitrate and sterilized at 121°C for 15 min and 1.2 bar. After cooling, PB medium was inoculated with approximately 106 CFU/mL of the selected strain and cultured at 20°C for 48 h. The pH of the PB medium was determined using a pH meter (Orion star, Thermo Scientific). Sampling was carried out at each fermentation time (0, 12, 24, 36, and 48 h) and 12 mL for the enzymatic assays to reduce nitrate to nitrite. The efficiency of the selected strain to reduce nitrate and produce nitrite was evaluated by determining their residual content in the PB medium. The nitrate and nitrite concentration of the culture supernatant was measured using a nitrate ion meter (Horiba Advanced Techno Co., Ltd., Tokyo, Japan) and nitrite high range portable photometer (Hanna Ins., Woonsocket, RI, USA), respectively. Reduced nitrate and produced nitrite were expressed as parts per million (ppm) of their initial PB medium. All residual nitrate and nitrite assays were carried out in duplicate and all treatments within a block were simultaneously analyzed to minimize the temporal variation in the assay.
Results and Discussion
It was reported that kimchi cabbage (1,740 mg/kg), lettuce (2,430 mg/kg), spinach (4,259 mg/kg), and radish (1,878 mg/kg) have the highest nitrate content in Korea (Chung et al., 2003). Microorganisms often adapt to their microenvironment and exhibit excellent properties. Therefore, vegetable-derived kimchi with a high nitrate content was harvested to isolate microorganisms with excellent nitrite resistance. Thousand strains of nitrite-resistant bacteria including Leuconostoc, Weissella, Lactobacillus, Pediococcus, and Staphylococcus sp. were isolated from various types of kimchi including, cabbage kimchi, spinach kimchi, leaf mustard kimchi, turnip kimchi, young radish kimchi, and cubed radish kimchi. Thousand types of isolates were cultured in MRS medium supplemented with nitrate (NaNO3), followed by a screening assay for nitrite (NaNO2) production. Twenty-four kimchi-fermenting microorganisms including Lactobacillus sakei (4 strains), L. plantarum (2 strains), L. brevis (2 strains), L. curvatus (2 strains), L. alimentarius (3 strains), Leuconostoc mesenteroides (3 strains), Leu. citreum (4 strains), Pediococcus inopinatus (3 strains), and Staphylococcus hominis (1 strain) with excellent nitrite and nitric oxide-producing potential were selected. Among nitrite-resistant isolates, nitrate content in the culture supernatant was measured, and isolate WiKim0113 was selected the strain with excellent nitrite and nitric oxide productivity (data not shown).
Isolate WiKim0113 was gram-positive, facultatively anaerobic, and formed grape-like clusters. It formed cream-colored, slightly elevated colonies on TSA at 30°C (data not shown). It was grown in NaCl (up to 9%; w/v) and at 10°C–40°C; its optimum growth was observed at 30°C at pH 4.0–10.0. Phenotypic characteristics and sugar utilization are summarized in Table 2.
Acid without gas was produced (weakly) with CHL suspension medium supplemented with the following sugars in the API 50CH gallery: galactose, D-glucose, D-fructose, N-acetyl glucosamine, maltose, lactose, saccharose, trehalose, melezitose, and D-turanose. Acid was not produced from the following sugars: glycerol, erythritol, D-arabinose, L-arabinose, ribose, D-xylose, L-xylose, adonitol, β-methyl-xyloside, D-mannose, L-sorbose, rhamnose, dulcitol, inositol, mannitol, sorbitol, α-methyl-D-mannoside, α-methyl-D-glucosamine, amygdaline, arbutine, aesculin, salicin, cellobiose, melibiose, inulin, D-raffinose, amidon, glycogen, xylitol, β-gentiobiose, D-lyxose, D-tagatose, D-fucose, L-fucose, D-arabitol, L-arabitol, gluconate, 2-ketogluconate, and 5-ketogluconate.
WiKim0113 produced high levels of nitrite; hence, this strain, with high nitrate reductase activity, was speculated to improve nitrate reduction to nitrite during fermentation. The 16S rRNA gene sequence of WiKim0113 was compared to those in the GenBank database via BLASTN, and a phylogenetic tree was constructed using the neighbor-joining method (Fig. 1). Close relationships in the phylogenetic tree facilitated subsequent identification of the bacterial species represented by 16S rRNA gene sequences. WiKim0113, with its superior nitrate reductase activity, was identified as a strain of Staphylococcus hominis subsp. hominis (98% sequence homology) upon 16S rRNA sequence analysis and was herein designated as S. hominis subsp. hominis WiKim0113.
S. hominis subsp. hominis WiKim0113 did not have hemolytic activity when grown in sheep blood agar. Hemolytic activity represents the safety of a culture starter (FAO and WHO, 2002). Herein, S. hominis subsp. hominis WiKim0113 exhibited non-proteolytic activity, whereas the positive control Bacillus sp. from kimchi exhibited proteolytic activity. The Bacillus sp. isolates were then characterized for protease production (data not shown).
Skim milk agar is commonly used to assess proteolysis by microorganisms capable of hydrolyzing casein. Proteolytic bacteria use protease to hydrolyze casein and form soluble nitrogenous compounds, characterized by a zone of clearance around colonies. It was reported that the activity of bioactive substances such as antibiotics and enzymes can be expressed in terms of the square of the diameter of the clear zone (Cooper, 1955). Psychrotrophs such as Pseudomonas sp. are strongly proteolytic and often responsible for spoilage of meat and dairy foods, thus resulting in a stale, bitter, or rancid flavor and smell. Therefore, protease-producing microorganisms are not suitable for the fermentation of meat products (Ercolini et al., 2009).
Enzyme activities of S. hominis subsp. hominis WiKim0113 are shown in Table 3. S. hominis subsp. hominis WiKim0113 displayed considerable alkaline phosphatase, esterase, trypsin, acid phosphatase, naphthol-AS-BI-phosphohydrolase, β-galactosidase, and α-glucosidase activity in the API ZYM system. The API ZYM system is a rapid semiquantitative procedure facilitating the detection of 19 enzymatic reactions. These data were harnessed for taxonomic classification and virulence evaluation of these organisms (Bascomb and Manafi, 1998). Bacterial β-galactosidase leads to the formation of galacto-oligosaccharides (GOS), which stimulate the growth and colonization of Bifidobacteria in the human intestine and suppress potentially harmful bacteria including Clostridium and Bacteroides spp. in the intestine (Sako et al., 1999). The results are similar to those of Staphylococcus xylosus with high nitrate reductase activity. It was reported strong positive reactions for alkaline phosphatase, esterase, esterase-lipase, acid phosphatase, and naphthol-AS-BI-phosphohydrolase in Staphylococcus xylosus in API ZYM assays (Foster et al., 1997). Staphylococcus aureus and novobiocin-sensitive coagulase-negative staphylococci produced acid and alkaline phosphatases, butyrate esterase, and caprylate esterase lipase. The color reaction of acid and alkaline phosphatases proceeded more rapidly with S. aureus (Humble et al., 1977). β-Glucuronidase and β-glucosidase were not detected in the API ZYM system of S. hominis subsp. hominis WiKim0113. None of microorganisms produced enzymes including β-glucuronidase, which stimulate colon cancer by converting pre-carcinogens into proximal carcinogens (Kim and Jin, 2001). Since the food industry requires careful assessment of the safety and usefulness of strains prior to their use in food, our results clearly indicate the suitability of S. hominis subsp. hominis WiKim0113 for its safety and utility as a culture starter (Parvez et al., 2006).
Agar diffusion method was used to carry out antibiotics susceptibility test. It was observed that S. hominis subsp. hominis WiKim0113 was highly sensitive to different classes of 14 antibiotics. The inhibitory zones were observed for ampicillin (21.67±0.58 mm), cefotetan (15.00±1.00 mm), chloramphenicol (31.00±1.00 mm), ciprofloxacin (34.67±1.15 mm), clindamycin (33.33±1.15 mm), doxycycline (35.67±1.15 mm), erythromycin (32.67±0.58 mm), gentamicin (34.00±1.73 mm), kanamycin (33.00±1.00 mm), penicillin G (19.00±1.00 mm), streptomycin (25.00±0.00 mm), tetracycline (35.33±0.58 mm), trimethoprim-sulfamethoxazole (35.00±0.00 mm), vancomycin (22.00±0.00 mm). S. hominis subsp. hominis WiKim0113 was sensitive to clinically relevant antibiotics. In addition, it appears to pose a lower risk for use in foods.
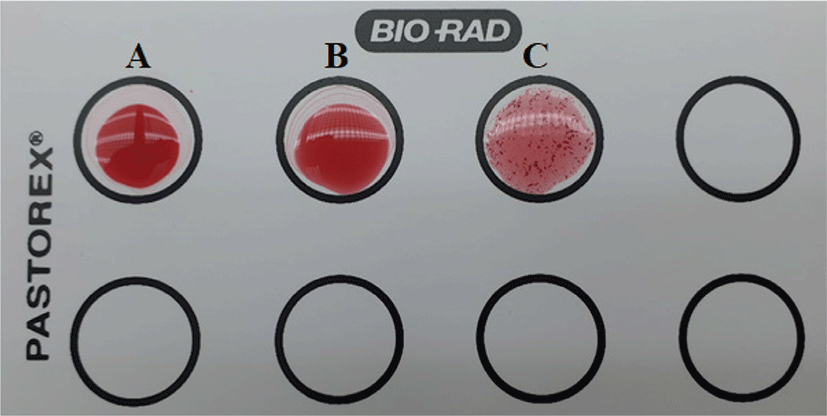
A comparative analysis for coagulase activity of Staphylococcus hominis subsp. hominis WiKim0113 by commercial Pastorex Staph-plus rapid agglutination test was shown in Fig. 2. Pastorex Staph showed negative (–) agglutination for S. hominis subsp. hominis, while strongly positive (+) agglutination was detected the presence of clumping factor on S. aureus ATCC25923. The sensitivity for Pastorex Staph-plus rapid agglutination test, consisting of a mixture of latex particles coated with fibrinogen and immunoglobulin G for the detection of bound coagulase (clumping factor) and protein A and latex particles sensitized with specific monoclonal antibodies to serotypes 5 and 8 capsular polysaccharides of Staphylococcus aureus, was higher than those of the other tests. It is known that the test kit can be used as a rapid reliable diagnostic test for identification of Staphylococcus aureus (Andriesse et al., 2011). It was previously reported that Staphylococcus hominis is a coagulase-negative member of the bacterial genus Staphylococcus, consisting of gram-positive, spherical cells in clusters and occurs very commonly as a harmless commensal on human (Weinstein et al., 1998).
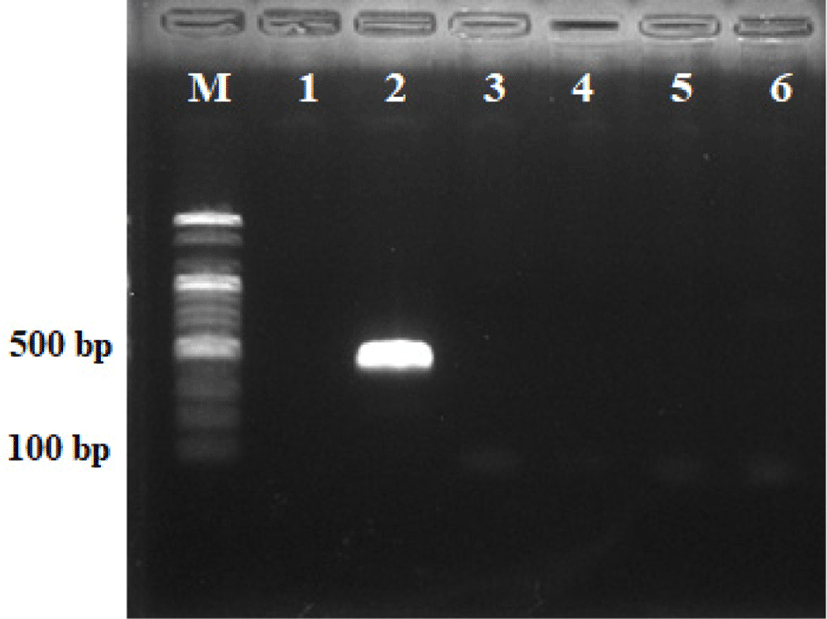
Staphylococcal enterotoxins (SEB and SEC) belong to a family of proteins of which immunologically distinct toxins and toxic shock syndrome toxin are recognized as virulence factors of Staphylococci. The SEs are recognized agents of the Staphylococcal food poisoning syndrome (Straub et al., 1999). Fig. 3 shows the results of molecular tests for the detection of genes encoding the staphylococcal enterotoxins (SEB and SEC), and toxic shock syndrome toxin-1 (TSST-1). Staphylococcus hominis WiKim0113 was negative for enterotoxin genes (SEB and SEC) and TSST-1 gene fragments, but Staphylococcus aureus ATCC25923 was positive for SEB genes. Staphylococcus aureus produces a spectrum of extracellular protein toxins and virulence factors which are thought to contribute to the pathogenicity of the organism. Members of the family cause toxic shock syndrome, while staphylococcal enterotoxin is the most common cause of food poisoning syndrome. Especially SEB is the most frequently observed enterotoxin in enterotoxigenic strains of S. aureus and ingested orally can cause severe gastrointestinal symptoms (Normannoa et al., 2005). Our results demonstrated the remarkable stability of Staphylococcus hominis WiKim0113, especially in strain negative for staphylococcal enterotoxins and TSST-1 genes.
Nitrite inhibits the growth of various bacterial strains. For processed meat products, nitrite is supplemented to prevent food poisoning caused by anaerobic bacteria including Clostridium botulinum, where residual nitrite levels are reportedly ≤20 ppm (Johnston et al., 1969). Hence, the antimicrobial activity of the culture supernatant of S. hominis subsp. hominis WiKim0113, was compared and analyzed relative to that of C. perfringens, which belongs to the same genus as C. botulinum.
Antagonistic activity was assayed by the agar well diffusion method. Herein, the culture supernatant of S. hominis subsp. hominis WiKim0113 displayed antibacterial activity (approximately 24 mm in diameter) against C. perfringens, which cause food poisoning symptoms (Fig. 4). However, MRS medium supplemented with nitrate was used as a negative control and did not exhibit antimicrobial activity. Nitrite is also effective against other foodborne pathogens including Bacillus cereus, Enterococcus faecalis, Listeria monocytogenes, E. coli O157:H7, and Staphylococcus aureus in meat products (Buchanan et al., 1989; Harrison et al., 1998; Lee et al., 2016; Redondo, 2011; Sameshim et al., 1997).

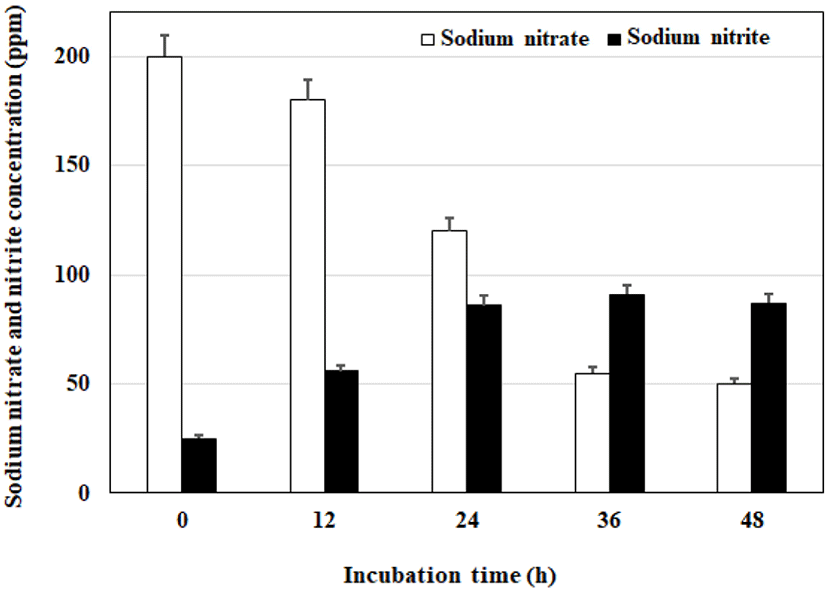
Staphylococcus hominis subsp. hominis WiKim0113 was cultured in PB medium with a similar composition to that in meat products, and nitrate reduction and nitrite production were assessed on the basis of residual nitrate and nitrite levels at 12-h intervals for 48 h. The results are expressed as ppm for nitrate reduction and nitrite production (Fig. 5). Herein, S. hominis subsp. hominis WiKim0113 exhibited 45.5% conversion of nitrate to nitrite with nitrate reduced to 25% after 36 h of culturing.
Nitrate reductase activities of Staphylococci is a common characteristic of several strains including S. simulans, S. sciuri, S. succinus subsp. casi, S. xylosus, and S. carnosus. Nitrite accumulates upon nitrate supplementation and inoculation of Staphylococci. Nitrate reductase activity is strain-dependent, ranging 41–796 nmol/min/mL. Several Staphylococcus strains with nitrite reductase activity ranging 4–42 nmol/min/mL were observed. It was reported that nitrate and nitrite reductase activities are greater in S. carnosus than in S. xylosus. (Gøtterup et al., 2007). Marked, highly effective NaNO2 production by the most efficient strain S. hominis subsp. hominis WiKim0113 and the abundant utilization of NaNO3 was achieved in 36 h cultures at 20°C and pH 6.3.
Conclusion
Most methods to determine suitable alternatives for synthetic nitrite in meat products depend on expensive, imported vegetable powder rich in nitrate or nitrite, or fermentation microorganisms in starter cultures, resulting in a high production cost and a foreign flavor. Hence, the unique alternative to nitrite based on kimchi-fermenting microorganisms and vegetables popularly consumed in Korea potentially provides a novel method to replace synthetic nitrite in accordance with consumer preferences and needs. Furthermore, source materials may be developed through this method. Herein, the substitution of synthetic nitrite using S. hominis subsp. hominis WiKim0113, having high nitrate reductase activity but no protein degradation activity and growing at low temperatures in the presence of nitrite, is potentially useful as a culture starter for fermented sausages.













