Introduction
The intestinal flora of an individual is composed of 100 trillion viable bacteria, representing 100 or more different bacterial species. These organisms live together in symbiotic or antagonistic relationships, growing on the food components ingested and biocomponents secreted into the alimentary tract by the host (Mitsuoka, 2000). Bifidobacteria are normal components of the intestinal flora throughout the life cycle. They are ubiquitous, endosymbiotic inhabitants of the gastrointestinal tract, vagina, and mouth. Bifidobacteria are the dominant species with Eubacterium, Clostridium, and Bacteroides in the colon flora, existing from 108 to 1011 per 1 g of colon contents (Mitsuoka, 1978).
Members of the genus Bifidobacterium are believed to exert various positive health benefits on their host. These beneficial effects include anticarcinogenic activity (Biffi et al., 1997; Rafter et al., 2007; Reddy and Rivenson, 1993; Rowland et al., 1998), modulation of immune response (Yasui and Ohwaki, 1991), improvement of the digestive system (Saavedra et al., 1994), and lowering incidences of necrotizing enterocolitis in preterm neonates (Patole et al., 2016). According to a recent report, there are some Bifidobacterium strains that can convert linoleic acid into bioactive conjugated linoleic acids (CLAs) (Coakley et al., 2003; O’Shea et al., 2012).
The term “CLA” refers to a group of positional and geometric isomers of essential fatty-acid linoleic acid (LA, cis9, cis12-C18:2) with conjugated double bonds, among which, the cis-9-,trans-11-ontadecadienoic acid (c9, t11 isomer) is approximately 80% of all possible CLA isomers (Pariza et al., 2001).
The conversion of linoleic acid to CLA takes place naturally in the rumen as part of the biohydrogenation process by the action of rumen bacteria such as Butyrivibrio fibrisolvens (Griinari and Bauman, 1999). Therefore, CLA is present as a natural component of ruminant meat as well as dairy products. There is increasing evidence of the potential health-promoting properties of CLA isomers, including antiatherosclerosis (Lee et al., 1994), anticarcinogenesis (Tian et al., 2011), enhancement of immunological function (Hayek et al., 1999), and reduction of body fat (Chaplin et al., 2015; Park et al., 1997; West et al., 1998).
Many probiotic bacteria, especially some bifidobacteria, have been reported to exhibit bioconversion activity from LA (linoleic acid) into CLA; however, this activity could be a strain-specific characteristic. From the screening of CLA production with the major species of bifidobacteria, it has been demonstrated that the most-efficient CLA producers belong to the species of B. breve (Barrett et al., 2007; Gorissen et al., 2010; Raimondi et al., 2016). In this study, a species-specific duplex polymerase chain reaction (PCR) technique for the selection of many strains belonging to the species of B. breve was developed, and this technique was successfully combined with a spectrophotometric assay for the rapid screening of probiotic candidates harboring CLA-producing activity.
Materials and Methods
First, traditional colonies from neonatal feces were propagated under anaerobic conditions in transoligosaccharide (TOS) agar (Yakult Honsha, Japan) at 37°C for 48 h (Thitaram et al., 2005).
Bifidobacterium isolates were activated by successive subculturing into a Man-Rogosa-Sharpe (MRS) (Difco, USA) broth supplemented with 0.05% L-cysteine·HCl and stored in 10% skim milk supplemented with glycerol in a –80°C deep freezer. As control strains, B. breve BB5 (Korea Yakult R & D Center) and B. breve LMC520 (Choi et al., 2008) were used for the CLA production activity.
Colonies grown on TOS agar were selected, and their genomic DNA was extracted with Lyse-N-Go (Pierce, USA) reagent according to the manufacturer’s protocol. The primers used in the duplex PCR were a genus-specific primer based on xfp genes, which encode fructose-6-phosphate phosphoketolase, and species-specific primers based on 16S rDNA from B. breve (Table 1).
| Primers | Description | Sequence (5’ to 3’) | Product size (bp) | References |
|---|---|---|---|---|
| GP-107 | Forward; 16S rDNA for B. breve | CCGGATGCTCCATCACAC | 250 | Matsuki et al. (2002) |
| GP-108 | Reverse; 16S rDNA for B. breve | ACAAAGTGCCTTGCTCCCT | Matsuki et al. (2002) | |
| GP-109 | Forward 1; xfp for Bifidobacterium | TGGCAGTCCAACAAGCTC | 923 | This study |
| GP-111 | Reverse ; xfp for Bifidobacterium | TAGGAGCTCCAGATGCCGTG | This study | |
| GP-110 | Forward 2; xfp for Bifidobacterium | CATCGACGGCAAGAAGACCG | 582 | This study |
| GP-111 | Reverse ; xfp for Bifidobacterium | TAGGAGCTCCAGATGCCGTG | This study | |
| GP-116 | Forward; 27F, universal for bacteria | AGAGTTTGATCCTGGCTCAG | 1,450 | |
| GP-120 | Reverse; 1492R, universal for bacteria | GGTTACCTTGTTACGACTT | ||
| GP-145 | Forward; BSH gene for B. breve | ATGTGCACTGGTGTCCGTTTC | 954 | This study |
| GP-146 | Reverse; BSH gene for B. breve | TCATCGGGCGACGCTGCTG | This study |
Colonies grown on TOS agar were selected, and the genomic DNA was extracted with Lyse-N-Go reagent, as recommended. Then, DNA was added in a mixture of 25 µL HotSTARTaq Master mix (Qiagen, USA), and a 1 µL forward and reverse primer set (GP116 & GP120) was used. The PCR product was purified using a PCR purification kit (Qiagen, USA) and identified by a Basic Local Alignment Search Tool (BLAST) search.
Primers used in the duplex PCR were genus-specific primers (GP109 & GP111) based on the xfp gene. The PCR product was ligated into the pGEM-T easy vector (Promega, USA). The ligation product was transformed E. coli JM109 (Takara, Japan) and grown in Luria-Bertani (LB) agar supplemented with 50 µg/mL of ampicillin and X-gal (LAX agar) plate at 37°C overnight. White colonies selected were inoculated in LB broth supplemented with 50 µg/mL of ampicillin and grown under 150 rpm at 37°C for 8 h. Plasmids from recombinant colonies were prepared by a miniprep kit (Qiagen, USA) and sequenced with a SP6 and T7 primer pair (Macrogen, Korea). The sequence was identified by BLAST search.
According to the method of Briczinski and Roberts (2006), 2 mL of the overnight cultured cell was centrifuged at 14,000 rpm for 10 min and resuspended with 600 µL of 100 mM Tris EDTA buffer (pH 7.6). 160 µL of sample was added to 40 µL of lysozyme (100 mg/mL) and 10 µL of proteinase K (20 mg/mL) and mixed with an equal volume of 1.6% InCert agarose (Bio-Rad Laboratories, USA) prepared in 0.1% SDS (International Biotechnologies, USA). Blended samples were dispended into disposable plug molds. The plugs were incubated in 0.5 M EDTA, 1% sarkosyl buffer (pH 9.0) supplemented by 2 mL of lysozyme (4 mg/mL) and mutanolysin (200 unit/mL) at 55°C for 90 min. After lysis, samples were cultured in fresh sarkosyl buffer supplemented with proteinase K (0.5 mg/mL) at 55°C for 1 h and washed in preheated distilled water at 50°C for 15 min. The plugs were washed three times with 10 mM EDTA buffer (pH 7.6) at 50°C for 15 min under 75 rpm in a shaking water bath. After lysis, the plugs were sliced at comb size and then transferred to 200 µL of a fresh restriction digest mixture containing 50 units of Xba I and incubated at 37°C for 2 h.
Electrophoresis was performed on 1% InCert agarose gel using a 0.5× TBE buffer (Bio-Rad Laboratories, USA). A lambda ladder (Bio-Rad Laboratories, USA) was included as a molecular size marker. An electrophoresis machine was operated using a CHEF DR-III (Bio-Rad Laboratories, USA). Operation times were increased linearly at 6 V/cm and 14°C. After electrophoresis, the gel was stained with ethidium bromide (0.4 mg/L; Promega, USA) for 1 h and then destained for 2 h with distilled water. The band pattern was confirmed using a UV transilluminator.
According to the method of Barrett et al. (2007), strains were incubated anaerobically in an mMRS broth containing linoleic acid (0.5 mg/mL) and 2% (wt/vol) Tween 80 at 37°C for 24 h. Following incubation, 1 mL of culture was centrifuged at 20,800×g for 1 min, the pellet was discarded, and the supernatant (0.5 mL) was mixed with 0.5 mL of hexane by vortexing for 2 min. 0.2 mL of the fatty acids extracted by vortexing the solution was mixed with 0.8 mL of methanol. The presence of CLA in the culture supernatant was assayed using a spectrophotometer (smart plus SP-1900PC, Youngwoo, Korea) at 233 nm.
According to the method of Jung et al. (2006), strains were incubated anaerobically in a mMRS broth containing linoleic acid (0.5 mg/mL) and 2% (wt/vol) Tween 80 at 37°C for 24 h. 0.5 mL of the sample was mixed with 20 μL of stock (8 mL of hexane, 100 mg of internal standard, C17:0) and 0.5 mL of hexane. After tilting for 10 min, the hexane layer was extracted by centrifuge. The hexane layer was dried under nitrogen gas at 60°C for 30 min.
Fatty acid was methylated in 1% HCl at 60°C for 30 min. Fatty-acid methyl esters were produced by adding 2 mL of saturated NaCl and 1 mL of hexane. The solvent was dried under nitrogen gas and concentrated with 100 µL of hexane. Methylated fatty acids were analyzed with a gas chromatographer (Varian STAR 3400, USA) equipped with an SP-2560 column and a flame ionization detector.
Results and Discussion
Twelve neonatal feces were plated on TOS agar and incubated under anaerobic conditions at 37°C for 48 h. Four hundred typical milky-white colonies were selected from agar plate and were subcultured in MRS broth. Isolated strains were stored in 10% skim-milk-supplemented glycerol in a –80°C deep freezer.
Conventional biochemical tests have some limitations in identifying specific species and discriminating a large number of isolates. Thus, in this study, duplex PCR that can simultaneously accomplish the rapid identification and the correct differentiation of B. breve was developed.
The PCR assay was conducted with genus-specific primers (Table 1; GP109 & GP111 and GP111 & GP110) for the genus Bifidobacterium, producing products of 923 bp and 582 bp, respectively. As a result of PCR with species-specific primers (GP107 & GP108), it was confirmed that 250 bp PCR product was obtained from only B. breve strains (Fig. 1).
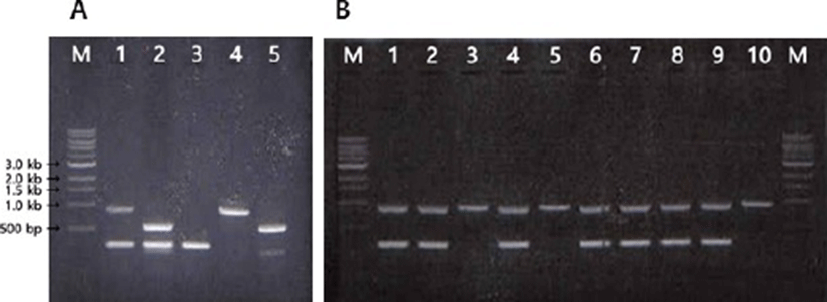
To increase the specificity of the PCR, a duplex PCR was conducted with multiplex primer sets, and two pairs of genus-specific primer (GP109 & GP111, GP145 & GP146) and a pair of species-specific primer (GP107 & GP108) were used. After the evaluation process, primer pairs (GP109 & GP111 and GP107 & GP108) producing products of 923 bp and 250 bp were selected because this set was better than the other sets producing products of 582 bp and 250 bp in terms of the band separation (Fig. 1A).
Through the duplex PCR assay developed in this study, we were able to confirm 36 strains B. breve from total 400 strains of Bifidobacterium isolated from new born infants. Some of the typical PCR band pattern for B. breve strains, which produce the PCR products of 923 bp and 250 bp are presented in Fig. 1B.
It is generally accepted that the predominant microorganisms in the colon of breast-fed babies are bifidobacteria, while the colons of bottle-fed babies contain various other bacteria. Mitsou et al. (2007) reported that Bifidobacterium longum and Bifidobacterium breve were the most frequently detected Bifidobacterium species in breast-fed infants.
In this study, B. breve was isolated from two infants among 12 infants. Only one strain among 40 strains from one person was identified as a B. breve, but 31 strains among 40 strains from another person were identified as B. breve, proving significant differences depending on the host.
As a result of sequencing the 16S rDNA of B. breve by duplex PCR, all strains had high homology with B. breve, indicating that all strains were B. breve. If the sequence of the 16S rRNA gene has more than 97% species sequence identity, they are considered to be the same species (Stackebarandt and Goebel, 1994). Bifidobacterium breve ATCC 15700T and commercial strain BB5 have 98% sequence identity, and B. breve ATCC 15700T and IB84 strain have 99% sequence identity. As a result of comparing the nucleotide sequence by clustal W to confirm the difference between the strains of isolated B. breve, all strains have the same 16S rDNA except BB5. Although it is difficult to identify the strain because of the high homology of the DNA sequence between genus Bifidobacterium, studies, such as those on multiple PCR (Kwon et al., 2005; Ventura et al., 2004) and DGGE (Satokari et al., 2001), have been conducted to identify Bifidobacterium, and it was found to be a convenient and accurate method for the identification of strain level.
PCRs were conducted with genus-specific primers, and the PCR products were cloned into a pGEM-T easy vector. As a result of comparing the sequence by clustal W, the IB sample corresponded with B. breve ATCC 15700T, and the BB5 sample and B. breve ATCC 15700T showed difference in six nucleotides.
Rosselló-Mora and Amman (2001) reported that the 16S rRNA gene sequence cannot distinguish clearly between difference species belonging to the same genus of bacteria. Berthoud et al. (2005) identified Bifidobacterium by the xfp gene and reported that the xfp gene sequencing is more accurate than the 16S rRNA gene sequencing, except for B. thermophilum, B. thermacidophilum, and B. boum. In this study, it was also confirmed that in addition to 16S rRNA gene sequencing, xfp gene sequencing could be used as a tool of identification for the isolated B. breve strains (data not shown).
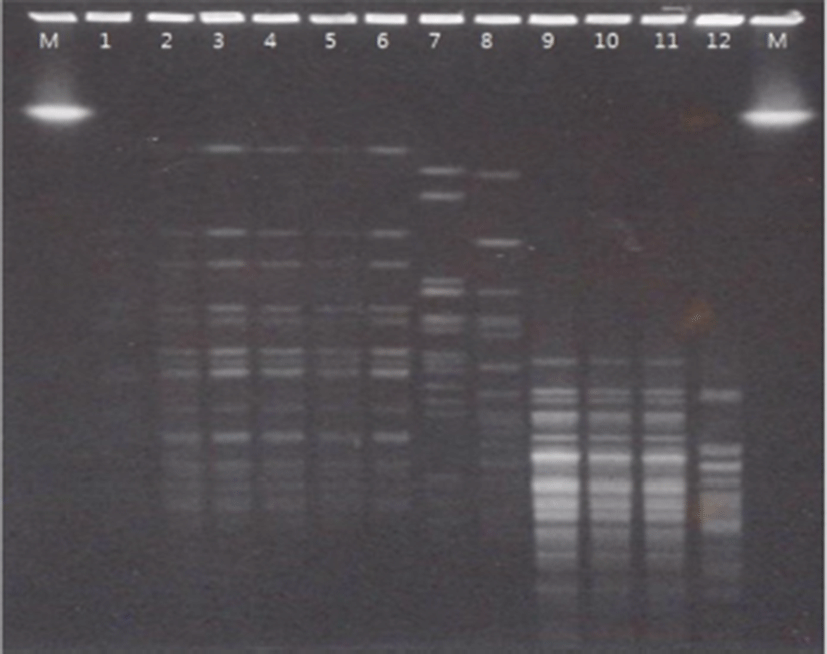
For the genotyping and genetic fingerprinting of the bacterial isolates, PFGE was applied using Xba I as restriction enzymes. As shown in Fig. 2, PFGE analysis indicated that there was high correlation between new isolated strains (lane 2 to lane 6) and different band pattern was observed from KCTC strains (land 8 to lane 11). Briczinski and Roberts (2006) reported that there is no difference between a PFGE protocol consuming 5–7 days and a rapid protocol that takes 24 h. In this study, PFGE was carried out using a rapid protocol, and it was confirmed that the band of the sample isolated from the same infants also showed the same patterns (Fig. 2).
The standard gas liquid chromatography-based screening process is laborious and time-consuming and can be a limiting factor when a large number of strains are being tested. In this study, a simple and straightforward spectrophotometric method was used for screening a large number of culture supernatants for the CLA production. Through the spectrophotometric assay, the present study confirmed that B. breve isolated from neonatal feces produces CLA, but three species of B. breve received from Korean Collection of Type Cultures (B. breve KCTC 3220, B. breve KCTC 3419, B. breve KCTC 5018) and B. breve 2003 do not produce CLA (Fig. 3).
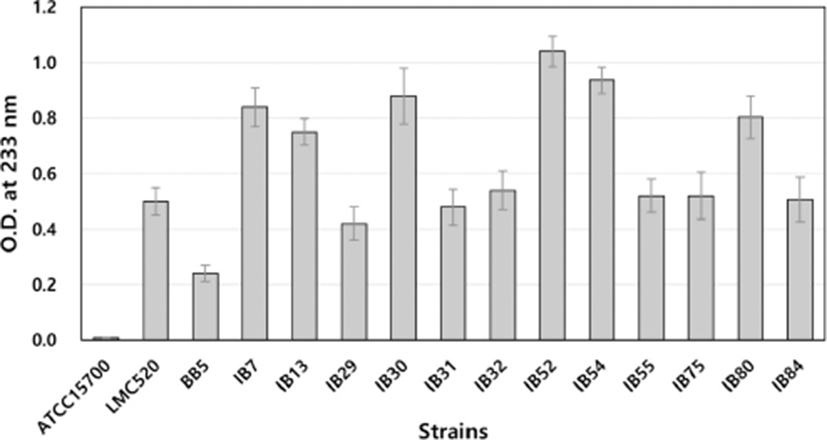
As a result of the spectrophotometric assay, CLA-producing ability was different depending on the strains tested, and it was observed that some of those isolates had higher CLA production activity than that of LMC520, which is one of the best CLA-producing strains (Choi et al., 2008).
Pariza and Yang (1999) reported that CLA detection is possible through the spectrophotometric assay, and Barrett et al. (2007) isolated and identified CLA-producing strains. In this study, the prescreening process was successful through the spectrophotometric assay, and the fatty acid analysis was carried out by GC for a more accurate quantitative analysis (Fig. 3).
Strains that had a high optical density value in the spectrophotometer assay were analyzed by GC to find excellent strains for the CLA production. First, the peak pattern of fatty-acid methyl ester (FAME) was analyzed using linoleic acid and conjugated methyl ester (Sigma, USA).
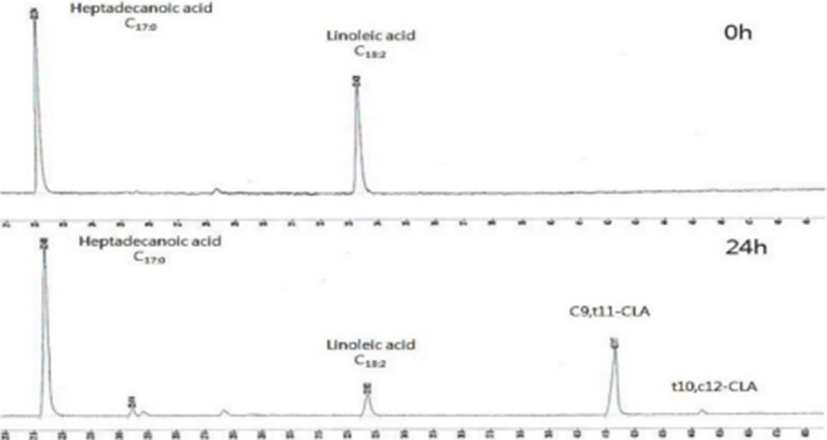
It was observed that linoleic acid was converted to CLA by B. breve isolated from neonatal feces through chromatograms (Fig. 4) with c9, t11-form as a major form of isomers of conjugated CLA. As shown in Fig. 5, conversion rates from LA into CLA was different depending on the strains. Among the isolated B. breve strains, IB52 and IB84 strains showed more than 65% conversion rate, which is comparable with LMC 520 strain (Choi et al., 2008). Many studies reported that free linoleic acid inhibits the growth of CLA-producing strains (Jiang et al., 1998; Kim et al., 2000; Rainio et al., 2001), and Tween 80 neutralizes the antibacterial activity and increases the solubility of the fatty acid in the liquid medium (Jiang et al., 1998; Rainio et al., 2001).
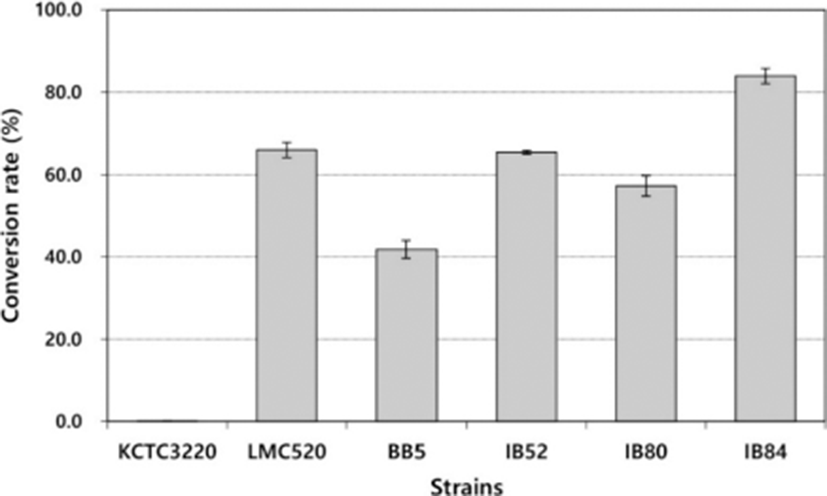
The study of the effect of the content of Tween 80 confirmed that the conversion rate of the medium containing 5% Tween 80 was higher than that of 2% Tween 80 (data not shown). The conversion rates of the isolates IB52, IB84, and LMC 520 were almost the same as that of LMC 520, and the conversion rate of IB84 was slightly higher than that of LMC 520.
Coakley et al. (2003) reported that B. breve and B. dentium have the highest productivity, comparing the conversation rate of CLA by Bifidobacterium isolated from neonatal feces, and Oh et al. (2003) confirmed that B. breve and B. pseudocatenulatum produce CLA. In this study, only B. breve had CLA-producing ability among 400 Bifidobacterium strains from neonatal feces.
Conclusion
Among the 400 strains identified out of 12 fecal isolates, 36 strains of B. breve were selected through duplex PCR. The screening study showed that IB52, IB80, and IB84 had CLA-producing ability converted from linoleic acid. In this study, using a duplex PCR for the selection of B. breve strains was a useful and precise method of selecting CLA-producing strains from human feces. Selected strains are expected to be widely used in materials of fermented milk products and health food. There are many studies that reported the efficacy of CLA-producing strains, including B. breve, through clinical trials. Further studies are necessary to clarify CLA-producing ability from linoleic acid by B. breve in the body.













