ARTICLE
Effects of a Novel p.A41P Mutation in the Swine Myogenic factor 5(MYF5) Gene on Protein Stabilizing, Muscle Fiber Characteristics and Meat Quality
Youn-Chul Ryu1,†, Eun-A Lee2,†, Han-Ha Chai3,†, Jong-Eun Park3, Jun-Mo Kim4,*
Author Information & Copyright ▼
1Division of Biotechnology, Sustainable Agriculture Research Institute, Jeju National University, Jeju 63243, Korea
2Division of Biotechnology, College of Life Sciences and Biotechnology, Korea University, Seoul 02841, Korea
3Division of Animal Genomics and Bioinformatics, National Institute of Animal Science, Rural Development Administration, Wanju 55365, Korea
4Department of Animal Science and Technology, Chung-Ang University, Anseong 17546, Korea
*Corresponding Author : Jun-Mo Kim Department of Animal Science and Technology, Chung-Ang University, Anseong 17546, Korea Tel: +82-31-670-3263 Fax: +82-31-675-3108 E-mail:
junmokim@cau.ac.kr
† These authors contributed equally to this study.
© Copyright 2018 Korean Society for Food Science of Animal Resources. This is an Open-Access article distributed under the terms of the
Creative Commons Attribution Non-Commercial License (http://creativecommons.org/licenses/by-nc/3.0/) which permits
unrestricted non-commercial use, distribution, and reproduction in any
medium, provided the original work is properly cited.
Received: Jun 08, 2018 ; Revised: Jun 20, 2018 ; Accepted: Jun 25, 2018
Published Online: Aug 31, 2018
Abstract
Myogenic factor 5 (MYF5) plays an important role in regulating skeletal muscle fiber characteristics, consequently affecting meat production and quality. We identified a novel p.A41P mutation in exon1 of the porcine MYF5 gene by direct sequencing. The mutation was predicted to be destabilizing in protein structure based on the resultant amino acid substitution. We estimated the significant substitution effect of p.A41P on the energy stabilization of Myf5 protein structure. Then, we demonstrated that the mutation in Yorkshire population significantly affected muscle fiber type I composition (p<0.05), loin-eye area of lean meat content (p<0.05) and filter-fluid uptake of meat quality (p<0.01). Furthermore, dominant effects significantly influenced total muscle fiber number (p<0.05). This study suggests that the novel p.A41P mutation in porcine MYF5 may be a valuable genetic marker to affect the muscle fiber characteristics and consequently improve meat production quality and quantity.
Keywords: Myogenic factor 5 (MYF5); single nucleotide polymorphism (SNP); muscle fiber characteristics; lean meat content; meat quality
Introduction
Myogenic regulatory factor (MRF) genes encode highly conserved basic helix-loop-helix proteins that control the embryonic muscle development process (Olson, 1990). Myogenic factor 5(MYF5) encodes a MRF named myogenic factor 5 (Atchley et al., 1994). Along with MYOD1, MYF5 expression is induced in myoblasts and is important to the regulation of myogenic proliferation and differentiation into myofibers (Montarras et al., 1991). Disruption in mice of the MYF5 locus, but not of the MYOD1, leads to a delayed and reduced myogenesis (Braun et al., 1992a). The porcine MYF5 was previously mapped to chromosome 5 (Soumillion et al., 1997), and it comprises 3 exons; 500, 76 and 191 bps long (Te Pas et al., 1999). This gene likely function in formation of muscle fiber characteristics, and has been considered a candidate gene for lean meat production and meat quality (da Silva Carmo et al., 2005; te Pas and Visscher, 1994). Therefore, the aim of the current study was to find the novel genetic marker for lean meat production and meat quality, according to functional validation via protein stabilizing changes and association analysis between polymorphism of porcine MYF5 gene and the related traits.
Materials and Methods
Identification of p.A41P mutation in MYF5
Direct sequencing analysis was performed to identify the novel non-synonymous single nucleotide polymorphism (nsSNP) mutations in the porcine MYF5 gene. Oligonucleotide primers for the sequencing analysis were designed with forward (5’-TGCGGTGGGATATGCTAATA-3’) and reverse primers (5’-CTCTGGTTGGGGTTAGTCGT-3’) based on published sequence data (GenBank ID. Y17154.1). A conventional polymerase chain reaction (PCR) amplification that produces a 600-bp fragment was conducted as follows: After heating at 95℃ for 10 min, then 35 cycles were adapted for denaturation at 95℃ for 1 min, annealing at 60℃ for 1 min, and polymerization at 72℃ for 1 min. The amplified PCR products were purified with QIAquick PCR purification kit (Qiagen, Inc., Venlo, Netherlands) and bidirectionally sequenced on an ABI 3730 automated sequencer with Big-Dye terminator cycle sequencing reagents (Applied Biosystems, Foster City, CA, USA).
Validations of p.A41P mutation effects on protein stabilizing and phenotypes
To investigate the substitution effect on the energy stabilizing of protein structure via nsSNP mutation, we used the molecular modeling package and protein design program in the Discovery Studio (DS) 4.0 (Accelrys Inc., San Diego, CA, USA) to estimate the amino acid substitution effect on model structure (Spassov and Yan, 2013).
The p.A41P mutation was genotyped on 429 Yorkshire pigs by the RFLP analysis along with the HhaI restriction enzyme. The Yorkshire population was chosen randomly from a single farm and slaughtered across an average of 188.9±20.45 days, following standard guidelines from the Korean grading service for animal products. Backfat thickness was measured at the 11th and last thoracic vertebrae. The mean of these 2 measurements was used as the backfat thickness value. The loin eye area was measured at the level of the last rib. Carcasses were chilled at 4℃ for 24 h, after which the longissimus dorsi (LD) muscle was obtained to evaluate muscle fiber characteristics and meat quality traits. As we previously described in details (Kim et al., 2009), muscle fibre characteristics were estimated using the myosin ATPase activities via histochemical analysis and meat quality traits were tested by pH values at 45 min (pH45min), drip loss, filter paper fluid uptake (FFU) and lightness (L*). The association analysis of the mutation with both sets of traits was conducted by the GLM procedure in SAS (ver. 9.3, SAS Institute) as following the model: yijklm=μ+Mi+Sj+Bk+b1Sdayl+eijklm, where yijklm denotes the observed traits, μ is the overall population mean, and Mi and Sj are the fixed effects of the ith genotype and jth sex. Bk is a random effect for the kth batch of slaughter, b1Sdayl is a covariance regression coefficient for the day of slaughter, and ejjklm is the random residual error. Multiple comparisons of the least-square means between genotypes were performed using the SAS PDIFF option with a Tukey-Kramer adjustment.
Results and Discussion
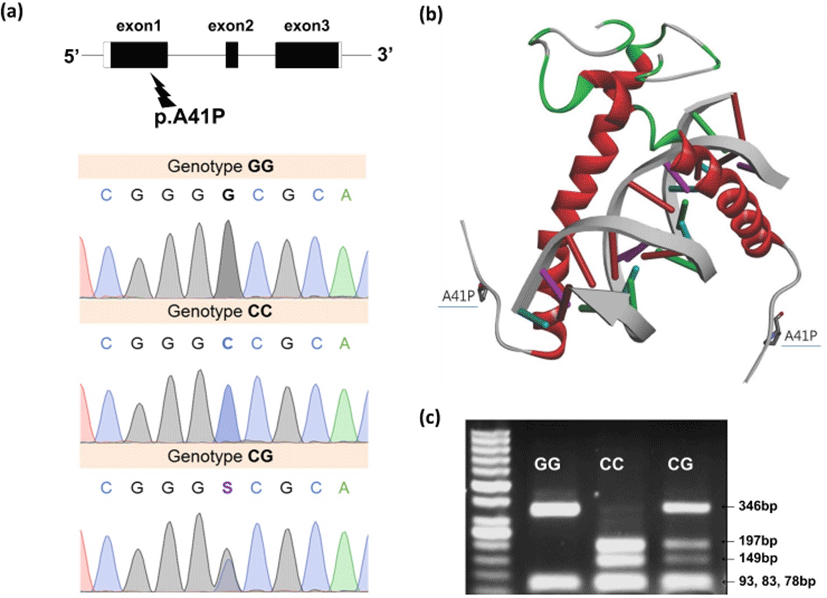
We identified a novel non-synonymous single nucleotide polymorphism (nsSNP, p.A41P via g.1121G>C) in the exon 1 region of Yorkshire pig MYF5 by direct sequencing (Fig. 1a). This amino acid substitution mutation (p.A41P) was located in the exposed loop of the Myf5 protein’s basic helix-loop-helix (bHLH) domain, rather than in the DNA-binding interface (Fig. 1b). Efficient DNA binding of Myf5 requires dimerization with another Myf5 (Winter et al., 1992). Therefore, the model structure for Myf5 bHLH domain-DNA binding complex was constructed upon a dimeric Myf5 complex. The substitution effect of p.A41P on the entropy term was predicted to be significantly destabilizing (ΔΔGmut>0.5 kcal/mol) via an increase of 2.18 kcal/mol in mutation energy compared with wild-type. Transcriptional activation via MYF5 requires activation of domains found in the amino- and carboxyl-terminal ends of the MYF5 peptide, along with the highly conserved bHLH domain encoded by exon 1 (Braun et al., 1992a; Braun et al., 1992b; Winter et al., 1992). Therefore, we supposed that the newly identified non-synonymous mutation in MYF5 gene may lead to destabilizing of Myf5 bHLH domain leading to structural change in DNA binding complex. It presumably could be a trigger of transcriptional activation of downstream target genes of MYF5 and consequently effect phenotypic changes.
Fig. 1.
(a) Identification of p.A41P mutation in the exon 1 region of MYF5 gene on chromosome 5 in Yorkshire pigs by direct sequencing: GG, homozygote for proline (P) substitution; CC, homozygote for alanine (A) wild-type; and CG, heterozygote. (b) Stereoview of model structure for Myf5 bHLH domain-DNA complex, drawn as a ribbon by using the molecular modeling package in the Discovery Studio (DS) 4.0 (Accelrys Inc., San Diego, CA, USA). (c) The 600 bp PCR product was digested with HhaI (New England Biolabs, MA, USA), resulting in products 345, 93, 83 and 78 bp (genotype GG).
The homozygote C allele (CC) resulted in product 197, 149, 93, 83 and 78 bp. The heterozygote (GC) resulted in 346, 197, 149, 93, 83 and 78 bp. The 93 bp, 83 and 78 bp fragments were shown as one band. The DNA fragments were separated on 3% agarose gels in 1× Tris-borate-EDTA buffer at 100 V for 30 min. MYF5, myogenic factor 5.
Download Original Figure
In this study, we used 429 Yorkshire pigs as a study population to validate the mutation effects on measured phenotypes such as muscle fiber characteristics and meat quality traits. The basic statistics (i.e., number of measurements per trait, means, standard deviations, minimum and maximum) for every measured trait were given in the Table 1. The p.A41P locus was genotyped for all the animals (Fig. 1c), and showed significant difference in their genotype frequencies from Hardy-Weinberg equilibrium (Table 2). Then, the association analysis revealed the significant effects of the p.A41P genotypes on the muscle fiber characteristics, lean meat production, and meat quality traits (Table 3). The p.A41P mutation was significantly associated with muscle fiber type I composition in both area and number (p<0.05). In mouse, MYF5 plays a role in the upregulation and activation of the developmental myosin heavy chain genes (Beylkin et al., 2006). Moreover, a previous study reported a SNP in porcine MYF5 has an influence on fast-twitch oxidative fiber contents of longissimus lumborum muscle (Klosowska et al., 2004). Therefore, it leads us to suggest that the novel nsSNP in exon 1 of MYF5 could have an effect on the formation of muscle fiber types.
Table 1.
Summary statistics for measured traits in 429 Yorkshire pigs
| Traits |
N |
Mean |
SD |
Min |
Max |
| Muscle fiber characteristics |
| Total fiber number (×103) |
429 |
1,165 |
260 |
523 |
2,159 |
| Fiber number per unit area (/mm2) |
429 |
242.0 |
34.9 |
149.0 |
368.0 |
| CSA of fibers (μm2) |
429 |
4,219 |
622 |
2,718 |
6,691 |
| Fiber number composition (%) |
| Type I |
429 |
9.23 |
3.98 |
1.23 |
30.00 |
| Type IIa |
429 |
13.97 |
5.17 |
0.85 |
37.91 |
| Type IIb |
429 |
76.80 |
6.51 |
48.01 |
92.12 |
| Fiber area composition (%) |
| Type I |
429 |
6.94 |
2.85 |
1.35 |
16.47 |
| Type IIa |
429 |
8.31 |
3.34 |
0.58 |
17.85 |
| Type IIb |
429 |
84.75 |
4.58 |
70.94 |
95.72 |
| Lean meat production |
| Loin-eye area (cm2) |
429 |
48.17 |
8.29 |
24.59 |
73.43 |
| Backfat thickness (mm) |
429 |
20.99 |
5.82 |
6.00 |
36.00 |
| Meat quality |
| pH45min |
428 |
6.13 |
0.28 |
5.34 |
6.94 |
| Drip loss (%) |
429 |
3.35 |
1.98 |
0.57 |
13.31 |
| Filter-fluid uptake (mg) |
429 |
28.19 |
15.83 |
5.10 |
99.30 |
| Lightness (L*) |
429 |
46.41 |
2.82 |
33.47 |
54.27 |
Download Excel Table
Table 2.
Genotype distribution of p.A41P mutation in 429 Yorkshire pigs
| Genotype count |
MAF |
Het |
χ2 |
| Total |
429 |
|
|
|
| CC |
29 |
0.331 |
0.527 |
15.41 |
| CG |
226 |
|
|
|
| GG |
174 |
|
|
|
Download Excel Table
Table 3.
Effects of p.A41P mutation in myogenic factor 5 (MYF5) on muscle fiber characteristics and economic traits in 429 Yorkshire pigs
| Traits |
p.A41P Genotype |
Additive |
Dominant |
Significance |
| CC (29) |
CG (226) |
GG (174) |
G |
A |
D |
| Muscle fiber characteristics |
| Total fiber number (×103) |
1,169±41.2 |
1,226±18.4 |
1,177±20.5 |
–8.906 |
105.2 |
|
ns |
|
| Fiber number per unit area (/mm2) |
234.6±6.36 |
242.5±2.85 |
241.6±3.17 |
–7.061 |
8.748 |
ns |
ns |
ns |
| CSA of fibers (μm2) |
4,392±114.2 |
4,215±51.1 |
4,217±56.9 |
174.5 |
–179.9 |
ns |
ns |
ns |
| Fiber number composition (%) |
| Type I |
11.03±0.76X |
9.17±0.34Y |
9.03±0.38Y |
2.006 |
–1.717 |
|
|
|
| Type IIa |
14.65±0.95 |
14.70±0.42 |
15.17±0.47 |
–0.515 |
–0.430 |
ns |
ns |
ns |
| Type IIb |
74.32±1.18 |
76.13±0.53 |
75.81±0.59 |
–1.490 |
2.145 |
ns |
ns |
ns |
| Fiber area composition (%) |
| Type I |
8.08±0.55X |
6.75±0.25Y |
6.76±0.27Y |
1.323 |
–1.330 |
|
|
|
| Type IIa |
8.84±0.61 |
8.77±0.27 |
9.09±0.31 |
–0.250 |
–0.396 |
ns |
ns |
ns |
| Type IIb |
83.08±0.84 |
84.48±0.37 |
84.15±0.42 |
–1.075 |
1.726 |
ns |
ns |
ns |
| Lean meat production |
| Loin–eye area (cm2) |
49.76±1.24XY |
50.59±0.55X |
48.68±0.62Y |
1.079 |
2.739 |
|
|
|
| Backfat thickness (mm) |
22.28±0.79 |
21.00±0.35 |
20.91±0.39 |
1.362 |
–1.189 |
ns |
ns |
ns |
| Meat quality |
| pH45min |
6.12±0.05 |
6.14±0.02 |
6.16±0.02 |
–0.037 |
–0.008 |
ns |
ns |
ns |
| Drip loss (%) |
3.56±0.37 |
3.33±0.17 |
3.02±0.19 |
0.545 |
0.087 |
ns |
ns |
ns |
| Filter–fluid uptake (mg) |
30.41±2.74X |
27.75±1.22X |
23.58±1.37Y |
6.837 |
1.500 |
|
|
ns |
| Lightness (L*) |
46.67±0.49 |
46.13±0.22 |
46.19±0.24 |
0.478 |
–0.598 |
ns |
ns |
ns |
Download Excel Table
Additionally, we observed that the p.A41P mutation was significantly associated with loin-eye area (p<0.05) and filter-fluid uptake (p<0.01). Variations in porcine MYF5 were reported to be associated with meat quality traits especially including moisture content of LD muscle and water holding capacity (da Silva Carmo et al., 2005; Liu et al., 2008; Liu et al., 2007). MYF5 gene is located at SSC5q25 (Čepica et al., 1999), and mapped near the drip loss quantitative trait loci (QTL) regions (Jennen et al., 2007). Moreover, another SNP in MYF5 has been reported to be associated with lean meat content (Verner et al., 2007). Our results showed that additive genetic effects were significant and in line with the results of genotype associations (p<0.05). Moreover, dominant effects significantly influenced total muscle fiber number (p<0.05), while the influence on muscle fiber type I composition and loin-eye area were trended to near significance (p<0.10). The number of muscle fibers at birth in piglets was regulated by MRF genes (Handel and Stickland, 1987), and low birth weight was associated with impaired pre-natal muscle development (Foxcroft et al., 2006). In addition, piglets with low birth weights had less lean meat content (Gondret et al., 2005; Paredes et al., 2013; Rehfeldt et al., 2008). Taken together, the novel p.A41P mutation in porcine MYF5 had impact on the muscle fiber formation and thus lean meat content.
Conclusion
Overall, the novel non-synonymous SNP (p.A41P via g.1121G>C) in the exon 1 region of MYF5 was predicted to destabilize the protein structure, and had impact on the muscle fiber formation and thus lean meat content. Based on these findings, we suggest that the p.A41P mutation could be a meaningful marker for muscle fiber regulation and to choice favourable pork when it applies as a potential target in the porcine breeding program.
Acknowledgements
This work was supported by “Cooperative Research Program for Agriculture Science and Technology Development” (Project No. PJ01187601), Rural Development Administration and a grant (715003-07) from the Research Center for Production Management and Technical Development for High Quality Livestock Products through Agriculture, Food and Rural Affairs Research Center Support Program, Ministry of Agriculture, Food and Rural Affairs, Korea.
References
Braun T, Bober E, Arnold HH. Inhibition of muscle differentiation by the adenovirus E1a protein: Repression of the transcriptional activating function of the HLH protein Myf-5. Genes Dev. 1992a; 6:888-902.


Braun T, Rudnicki MA, Arnold HH, Jaenisch R. Targeted inactivation of the muscle regulatory gene Myf-5 results in abnormal rib development and perinatal death. Cell. 1992b; 71:369-382.


Čepica S, Yerle M, Stratil A, Schröffel J, Redl B. Regional localization of porcine MYOD1, MYF5, LEP, UCP3 and LCN1 genes. Anim Genet. 1999; 30:462-478.


da Silva Carmo FM, Guimarães SEF, Lopes PS, Pires AV, Guimarães MFM, da Silva MVGB, Schierholt AS, de Moraese Silva K, de Miranda Gomide LA. Association of MYF5 gene allelic variants with production traits in pigs. Genet Mol Biol. 2005; 28:363-369.


Foxcroft GR, Dixon WT, Novak S, Putman CT, Town SC, Vinsky MDA. The biological basis for prenatal programming of postnatal performance in pigs. J Anim Sci. 2006; 84:E105-E112.


Gondret F, Lefaucheur L, Louveau I, Lebret B, Pichodo X, Le Cozler Y. Influence of piglet birth weight on postnatal growth performance, tissue lipogenic capacity and muscle histological traits at market weight. Livest Prod Sci. 2005; 93:137-146.


Handel SE, Stickland NC. Muscle cellularity and birth weight. Anim Sci. 1987; 44:311-317.


Klosowska D, Kuryl J, Elminowska-Wenda G, Kapelanski W, Walasik K, Pierzchala M, Cieslak D, Bogucka J. A relationship between the PCR-RFLP polymorphism in porcine MYOG, MYOD1 and MYF5 genes and microstructural characteristics of m. longissimus lumborum in Pietrain×(Polish Large White×Polish Landrace) crosses. Czech J Anim Sci. 2004; 49(3):99-107.

Liu M, Peng J, Xu D, Zheng R, Li J, Zuo B, Lei M, Xiong Y, Deng C, Jiang S. Associations of MYF5 gene polymorphisms with meat quality traits in different domestic pig (Sus scrofa) populations. Genet Mol Biol. 2007; 30:370-374.


Olson EN. MyoD family: A paradigm for development?. Genes Dev. 1990; 4:1454-1461.


Soumillion A, Vergouwe M, Erkens J, te Pas M, Rettenberger G, Lenstra J. Assignment of the porcine loci for MYOD1 to chromosome 2 and MYF5 to chromosome 5. Anim Genet. 1997; 28:37-38.

te Pas M, Harders F, Soumillion A, Born L, Buist W. Genetic variation at the porcine MYF-5 gene locus. Lack of association with meat production traits. Mamm Genome. 1999; 10:123-127.