Introduction
Salmonella is a major zoonotic, foodborne pathogen that asymptomatically colonizes the intestinal tract of poultry and represents the leading cause of poultry-associated outbreaks in Europe, accounting for over 41.3% of reported cases (European Food Safety Authority [EFSA] and European Centre for Disease Prevention and Control [ECDC], 2023). Colonized poultry often act as silent reservoirs and vehicles, continuously shedding Salmonella into the farm environment and feed system (Thorns, 2000). It facilitates horizontal transmission within flocks and increases the risk of carcass contamination post-slaughter. Notably, while only 13% of broiler flocks were colonized at slaughter, 55% of broiler carcasses were contaminated with Salmonella after processing (Rasschaert et al., 2008). Although various sanitary interventions, including carcass rinsing, chilling, and surface decontamination, are implemented during slaughter and processing to reduce microbial loads (Micciche et al., 2018), they are often insufficient to mitigate contamination from intestinal colonization. These findings highlight the need for effective control strategies during poultry husbandry to prevent downstream contamination and dissemination throughout the processing chain.
Antibiotics, such as tetracyclines, sulfonamides, aminoglycosides, and macrolides, have been administered with feed or drinking water at sub-therapeutic doses to control Salmonella in poultry husbandry (Parveen et al., 2007). However, this application of antibiotics as feed additives has contributed to the emergence and spread of antimicrobial-resistant (AMR) Salmonella strains throughout poultry production systems. In a previous study, over 75% of Salmonella strains isolated from poultry in Korea were resistant to ampicillin, cefotaxime, and tetracycline (National Institute of Health [NIH], 2025). Furthermore, the use of pharmaceutical products to promote rapid growth and maintain animal health in poultry husbandry has resulted in the accumulation of toxic and harmful residues in the products, posing risks to consumer health (Mund et al., 2017). Due to these concerns, the use of antibiotics in feed has been banned in many countries, including the EU, leading to the adoption of alternative feed additives such as organic acids, probiotics, and essential oils (Logue et al., 2024). However, these alternative feed additives often lack target specificity, contributing to the inconsistent efficacy against Salmonella (Kerek et al., 2023; Naeem and Bourassa, 2024). Therefore, the need for safe and selective alternatives has led to growing interest in bacteriophage (phage)-based feed additives.
Lytic phages are viruses that specifically infect and lyse bacterial cells, offering high specificity, self-replication, natural abundance, and excellent stability (Kim et al., 2023). These characteristics have led to the commercialization of several phage-based products, including SalmoFreshTM, SalmonellexTM, and PhageGuardTM. However, these commercial products have been predominantly applied to reduce Salmonella contamination of poultry carcasses (Micreos Food Safety, 2021) and poultry products (Hagens et al., 2018; Sukumaran et al., 2016). More recently, phage application in poultry has expanded from post-slaughter treatment to use as a feed additive during poultry husbandry. A recent study demonstrated that ad libitum administration of two lytic phages, SPFM10 and SPFM14, significantly reduced Salmonella colonization in broiler chickens after 42 days (Thanki et al., 2023). To date, only one phage-based product (Bafasal®, Proteon Pharmaceuticals, Łódź, Poland), a phage cocktail targeting Salmonella Gallinarum and Salmonella Enteritidis, has been developed as a feed additive for preventive or metaphylactic use during the husbandry phase (Clavijo et al., 2019; EFSA Panel on Additives and Products or Substances used in Animal Feed et al., 2024; Pelyuntha et al., 2022).
Here, Salmonella Typhimurium phage vB_StyS_KFSST1, previously isolated from poultry processing wastewater, is proposed as a new, potential biocontrol candidate for a feed additive. This phage exhibited excellent temperature stability and acid tolerance (Choi et al., 2020), making it suitable for feed formulation and combination treatment with other alternatives such as organic acids or probiotics. Based on the EFSA under Regulation (EC) No. 1831/2003, the commercial phage-based feed additives should provide information regarding safety, host range, in vitro and in vivo biocontrol efficacy, and storage stability (EFSA Panel on Additives and Products or Substances used in Animal Feed et al., 2021; EFSA Panel on Additives and Products or Substances used in Animal Feed et al., 2024). Since the previous study has demonstrated the physical stability of vB_StyS_KFSST1, the present study focuses on its functional and genomic features to evaluate the suitability of the phage for use as a feed additive targeting Salmonella serovars. Specifically, this study aims to evaluate its lytic activity and in vitro efficacy against various Salmonella serotypes, and to provide its genome features to confirm the absence of undesirable genes, including those related to lysogeny, antimicrobial resistance, and virulence.
Materials and Methods
A total of 17 Salmonella strains were used in this study (Table 1), comprising 11 reference strains obtained from the American Type Culture Collection (ATCC) and the National Culture Collection for Pathogens (NCCP), and six Salmonella isolates previously recovered from fresh produce and agricultural environments (Choe et al., 2023). These 6 Salmonella isolates were previously whole-genome sequenced at Max Rubner-Institut (MRI) at the Department of Microbiology and Biotechnology in Kiel, Germany (Kim et al., 2025a), and identified as S. Typhimurium (Salmonellaenterica GOVDG-1, S. enterica GORGM-1, and S. enterica PLGS-1), S. I 4,[5],12:i:- (S. enterica PSGS-1), S. Kentucky (S. enterica PSCD-1), and S. Montevideo (S. enterica CMCD-1; Kim et al., 2025b). Genome sequences of six phage-susceptible strains, such as S. Typhimurium ATCC 13311, S. Typhimurium ATCC 14028, S. Enteritidis ATCC 13076, Salmonella enterica GOVDG-1, Salmonella enterica GORGM-1, and Salmonella enterica PLGS-1, were retrieved from the National Center for Biotechnology Information (NCBI) GenBank database under accession numbers NZCP009102.1, CP043907.1, NZLSHA01000001.1, JBNDEH000000000, JBNDEL000000000, and JBNDEI000000000, respectively.
2) EOP≥0.50, strong lytic capacity; 0.01≤EOP<0.50, intermediate lytic capacity. EOP<0.01, weak lytic capacity.
3) These environmental Salmonella isolates were previously described by Choe et al. (2023), and their serotypes were predicted based on whole genome sequencing as S. Typhimurium (GOVDG-1, GORGM-1, and PLGS-1), S. I 4,[5],12:i:- (PSGS-1), S. Kentucky (PSCD-1), and S. Montevideo (CMCD-1; Kim et al., 2025b).
vB_StyS_KFSST1 was previously isolated from the rinsing water of the poultry processing facility (Orpum, Sangju, Korea), using S. Typhimurium ATCC 13311 as the indicator host strain (Choi et al., 2020). For high-titer propagation, host culture was prepared by inoculating 1% (v/v) overnight culture into 3 mL of modified nutrient broth (0.15 g/L CaCl2, 0.05 g/L MnSO4, 0.2 g/L MgSO4, 5 g/L NaCl, and 8 g/L nutrient broth) and incubating it at 37°C with vigorous shaking until reaching the logarithmic growth phase. The phage suspension was then added at a multiplicity of infection (MOI) of 1, followed by incubation under the same conditions for phage proliferation. After incubation, the culture was centrifuged at 4,000×g for 10 min, and the supernatant was filtered through a 0.22-μm pore-size filter (GVS, Sanford, ME, USA). This propagation process was scaled up by gradually increasing the culture volume and repeating the same procedure described above. The propagated phage, with a final titer of approximately 10–11 Log PFU/mL, was purified via polyethylene glycol precipitation, CsCl density-gradient ultracentrifugation, and subsequent dialysis in SM buffer, as previously described (Kim et al., 2021). The purified phage stock was finally stored in a glass vial at 4°C prior to use.
Each strain was cultivated in tryptic soy broth (TSB; DifcoTM, Detroit, MI, USA) at 37°C for 12 h. A 200 μL aliquot of each overnight culture was mixed with 4 mL of 0.4% TA soft agar and overlaid onto TSA plates. Ten microliters of phage suspension (8 Log PFU/mL) were spotted onto the surface of the bacterial lawns. After 16 h incubation at 37°C, the formation of a single plaque was confirmed to determine the lytic activity of vB_StyS_KFSST1 against the tested bacterial strains. Once plaque formation was confirmed, efficiency-of-plating (EOP) of the phage was determined using plaque assay (Kim et al., 2023). EOP is calculated by dividing the phage titer on the tested bacterial strain by the phage titer on the indicator host strain.
S. Enteritidis ATCC 13076 and S. Typhimurium ATCC 13311 were used as representative hosts to analyze infection kinetics. Each strain was cultured in TSB at 37°C for 16 h, and the overnight cultures were diluted 1:100 (v/v) in fresh TSB. For the infection kinetics analysis, 100 μL of the diluted bacterial culture and 100 μL of phage suspension were added into each well of a 96-well microplate to achieve a MOI of 1. The microplate was incubated at 37°C for 12 h, and bacterial growth was then monitored by measuring optical density at 640 nm (OD640) using a microplate reader (Synergy H1, BioTek, Charlotte, VT, USA). All measurements were performed in triplicate.
Genomic DNA of vB_StyS_KFSST1 was extracted using Phage DNA Isolation Kit (Norgen Biotek, Thorold, ON, Canada) according to the manufacturer’s instruction. The extracted DNA was then purified using AMPure XP beads (Beckman Coulter, Brea, CA, USA). DNA quality and concentration were assessed using NanoDrop (Peqlab, Erlangen, Germany) and a Qubit 3.0 fluorometer (Thermo Fisher Scientific, Wesel, Germany). DNA library preparation was performed using the ligation sequencing kit with native barcoding (SQK-NBD114.96, Oxford Nanopore Technologies, Oxford, UK) and sequencing was carried out on a PromethION 2 Solo sequencing device using an R10.4.1 flow cell (Oxford Nanopore Technologies). Raw signal data in POD5 format were basecalled and demultiplexed using the Dorado software (v. 0.9.5). The raw sequence data in FASTQ format were filtered for quality control using the fastplong pipeline (v. 0.2.2; parameter: minlength 500 and Q 15; Chen, 2023). The de novo assembly was subsequently conducted using the Flye (v. 2.9.5) with the --nano-corr parameter (Kolmogorov et al., 2019). After genome assembly, the quality of the genome sequence was assessed using the QUAST pipeline (Mikheenko et al., 2018). The assembled genome in FASTA format was subjected to further bioinformatic analyses.
Annotation of the phage genome was conducted using BV-BRC (Olson et al., 2023) and Pharokka pipeline (v1.7.0; Bouras et al., 2023). To evaluate the safety of vB_StyS_KFSST1, both the annotated genome and bacterial genomes of phage-susceptible strains were screened for antibiotic resistance genes, virulence factors, and prophage regions using ResFinder 4.1, the Virulence Factor Database (VFDB), and PHASTEST (Wishart et al., 2023), respectively. ResFinder 4.1 was used to determine the presence of acquired antibiotic resistance genes with 80% sequence similarity (Bortolaia et al., 2020), while VFDB was applied to detect known virulence factors associated with Salmonella spp. (Liu et al., 2022). The phage lifestyle was classified using PhageAI platform (https://phage.ai/). For phylogenetic and taxonomical analyses, the average nucleotide identity (ANI) between vB_StyS_KFSST1 and its close relatives was calculated using the FastANI pipeline (v1.33; Jain et al., 2018) with default parameters. Additionally, complete genome sequence based phylogenetic analysis was performed using Virus Classification and Tree Building Online Resource (VICTOR) with the d0 formula (Meier-Kolthoff and Göker, 2017), and its output file was uploaded to iTOL (https://itol.embl.de) for visualization of the phylogenetic tree.
Host range, EOP analysis, and infection kinetics of the phage were conducted in triplicates, and data were expressed as the mean±SD. Statistical analyses were performed using GraphPad Prism and InStat V.9 (GraphPad, San Diego, CA, USA). Student’s paired t-test and one-way analysis of variance (ANOVA) were used to compare data between and among groups, respectively, at p values of <0.05.
Results and Discussion
The host range of vB_StyS_KFSST1 (Table 1) was evaluated against 10 Salmonella serotypes with EOP analysis, since it had already been assessed against 39 major foodborne pathogens, including 8 Salmonella serotypes (Choi et al., 2020). vB_StyS_KFSST1 exhibited lytic activity exclusively against S. Enteritidis and S. Typhimurium, lysing all tested strains within these two serotypes, including three reference strains and three environmental isolates (GOVDG-1, GORGM, and PLGS). Additionally, the phage showed high EOP values (≥0.98) for all six lysed strains (Table 1). These results indicate that vB_StyS_KFSST1 possesses dual serotype-specific lytic activity with high efficiency against S. Enteritidis and S. Typhimurium.
Infection kinetics of the phage were further assessed against the representative host strains of S. Enteritidis ATCC 13076 and S. Typhimurium ATCC 13311 (Fig. 1). With both serotypes, absorbance began to decline rapidly from 1 h after phage infection, in contrast to the phage-free control. The sharp reduction in absorbance indicated early phage adsorption and initiation of bacterial lysis (Shao and Wang, 2008). After a gradual decrease during the first 3h, the growth inhibition was sustained until 12 h (Fig. 1). No notable recovery in bacterial growth was observed for both strains within the experimental period. These findings demonstrate that vB_StyS_KFSST1 effectively infected and controlled S. Enteritidis and S. Typhimurium, showing comparable and sustained lytic activity against both serotypes.
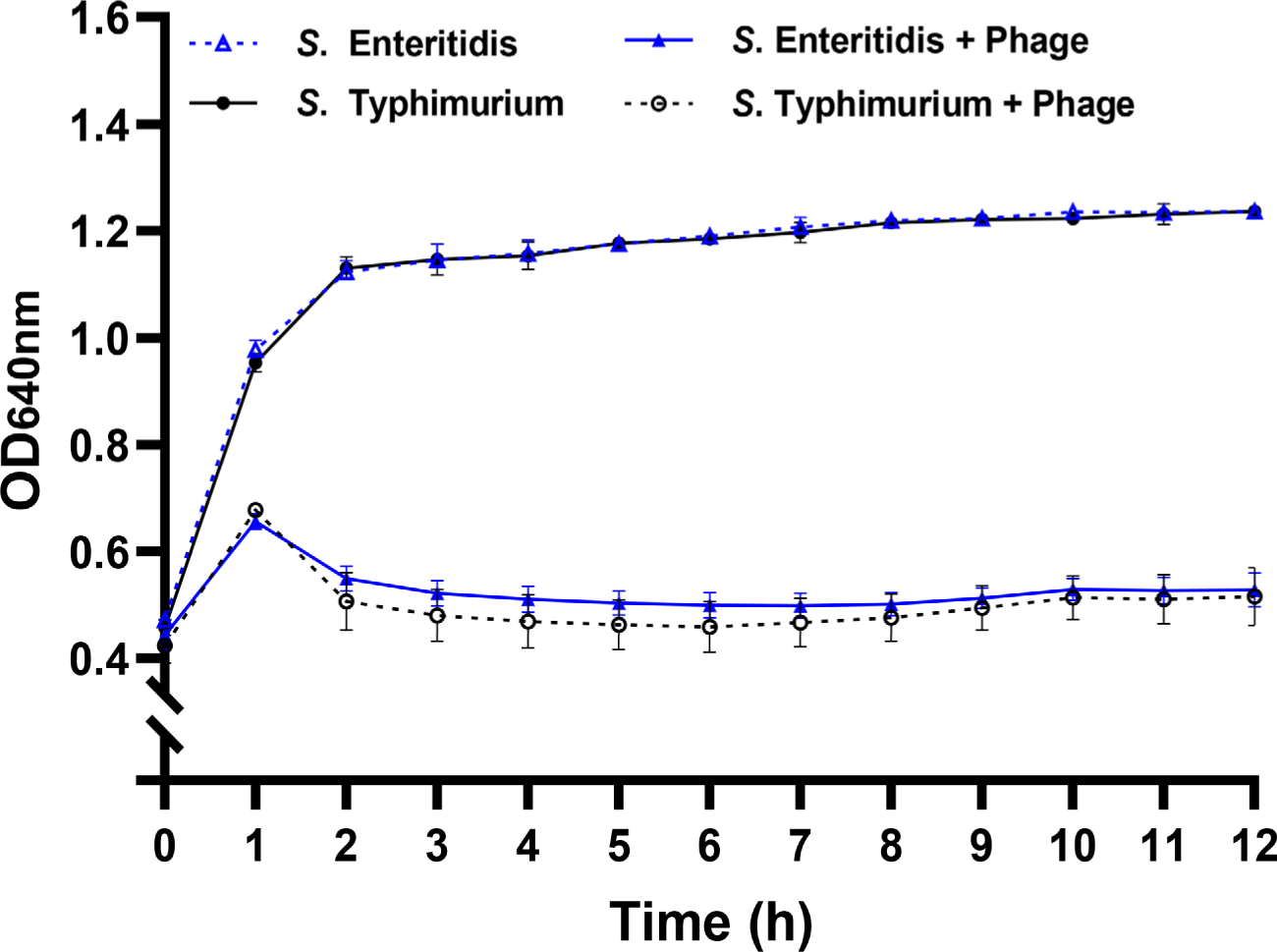
Similar to our phage, two Salmonella phages, L223 (Khan et al., 2024) and vB_Sen-TO17 (Kosznik-Kwaśnicka et al., 2022), also showed dual serotype-specific lytic activity against both S. Enteritidis and S. Typhimurium. However, these phages required at least ~3 h to initiate detectable growth inhibition, whereas vB_StyS_KFSST1 reduced bacterial growth within 1 h of phage infection. Compared to these studies, Salmonella phage SHWT1 showed the broader host range against a wider panel of Salmonella serotypes, including Derby, Enteritidis, Gallinarum, London, Pullorum, Typhi, and Typhimurium (Tao et al., 2021). However, its lytic activity was not sustained, as regrowth of host strains was observed after 2 h of phage infection, indicating incomplete antibacterial efficacy. Another previous study of phage phiSalP219 showed that this phage exhibited lytic activity against four Salmonella serotypes (Enteritidis, Gallinarum, Paratyphi, and Typhimurium), but also reported partial recovery of bacterial growth during the later stages of phage infection (Jaglan et al., 2024). In contrast, vB_StyS_KFSST1 achieved a rapid and maintained suppression of S. Enteritidis and S. Typhimurium without regrowth, consistent with its high EOP. Moreover, since Bafasal®, the first EFSA-approved phage product, is specifically targeted S. Gallinarum and S. Enteritidis, the application of vB_StyS_KFSST1 can expand the phage-based control strategy by covering S. Typhimurium, one of the most prevalent serotypes causing poultry-associated salmonellosis (Karabasanavar et al., 2020). These characteristics highlight the potential of vB_StyS_KFSST1 as a novel candidate for a feed additive for improving Salmonella control in poultry husbandry.
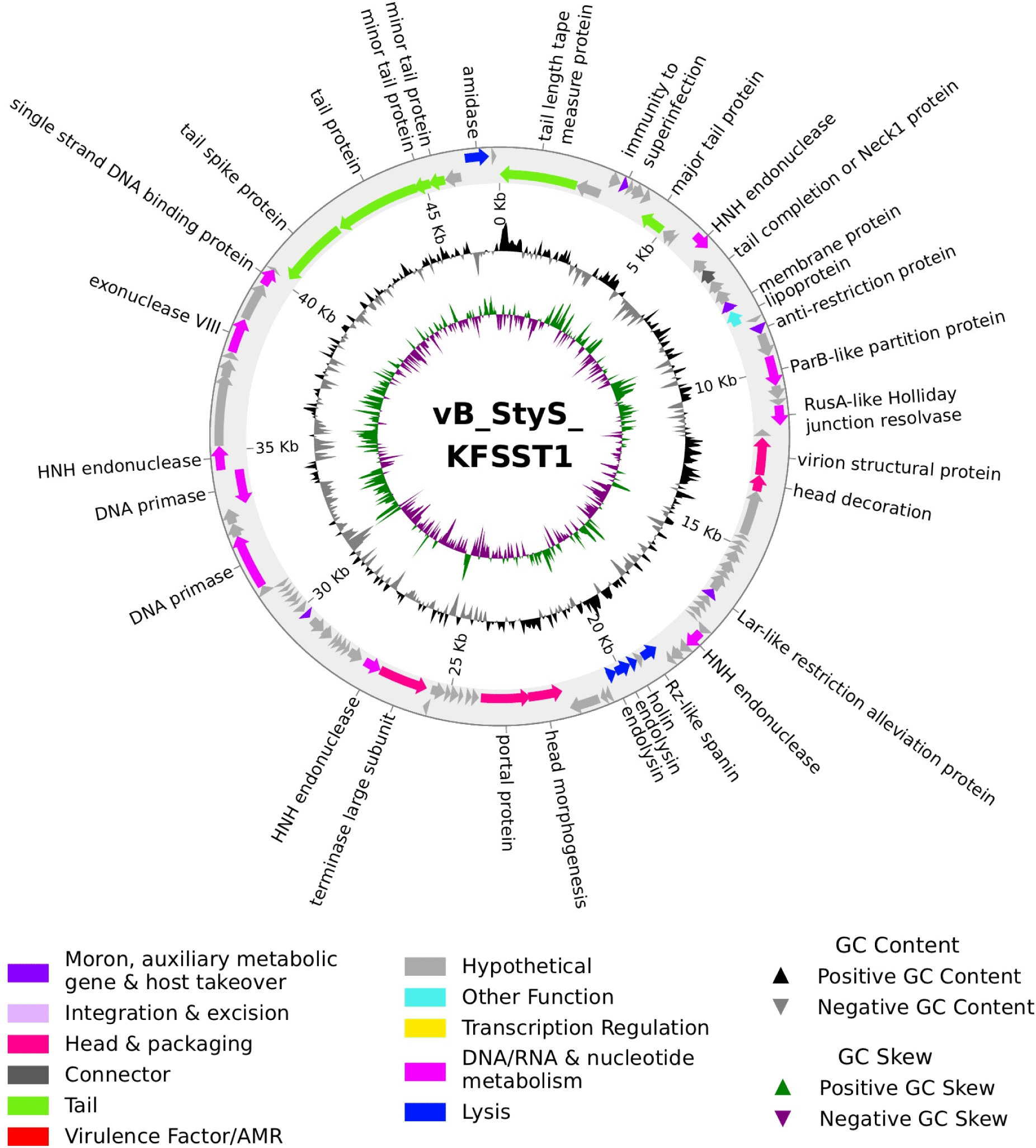
The complete genome of vB_StyS_KFSST1 consisted of double-stranded DNA with a total length of 47,149 bp and a mol% GC content of 45.74% (Fig. 2). The phage genome encodes 98 open reading frames (ORFs) and 2 tRNAs. With respect to start codon usage, the majority of predicted ORFs initiated with AUG (96.94%), while UUG and GUG accounted for 2.04% and 1.02%, respectively. Among the 98 ORFs, the function of only 33 ORFs could be predicted and categorized into six groups, including phage structure, DNA packaging, host lysis, nucleotide metabolism and replication, phage assembly, and additional functions (Table 2). The largest proportion of the functional ORFs were associated with structural components, such as phage tail, phage head, connector, and packaging proteins (Fig. 2). The remaining 65 ORFs were annotated as hypothetical proteins with unknown functions (Fig. 2). Notably, no integrase, repressors, or recombinase genes were detected, indicating that vB_StyS_KFSST1 is a strictly virulent phage. PHASTEST analysis additionally confirmed the absence of intact and incomplete prophage regions. Consistently, PhageAI predicted a virulent lifestyle with a 99.77% probability, further supporting the lytic nature and genetic stability of vB_StyS_KFSST1.
To determine the genomic features of vB_StyS_KFSST1, the presence and distribution of AMR genes and virulence factors were screened with an 80% identity threshold, together with a comparative analysis with the genomes of phage-susceptible Salmonellas trains. Several AMR genes were detected in the genomes of Salmonella host strains. These included aminoglycoside resistance genes [aac(6')-Iaa and aad(6')-ly], beta-lactam resistance genes (blaTEM-1B, and ampH), sulfonamide resistance gene (sul2), tetracycline resistance gene (tetA), and various multidrug efflux pump-related genes (Fig. 3A). In contrast, no AMR genes were detected on the genome of vB_StyS_KFSST1.
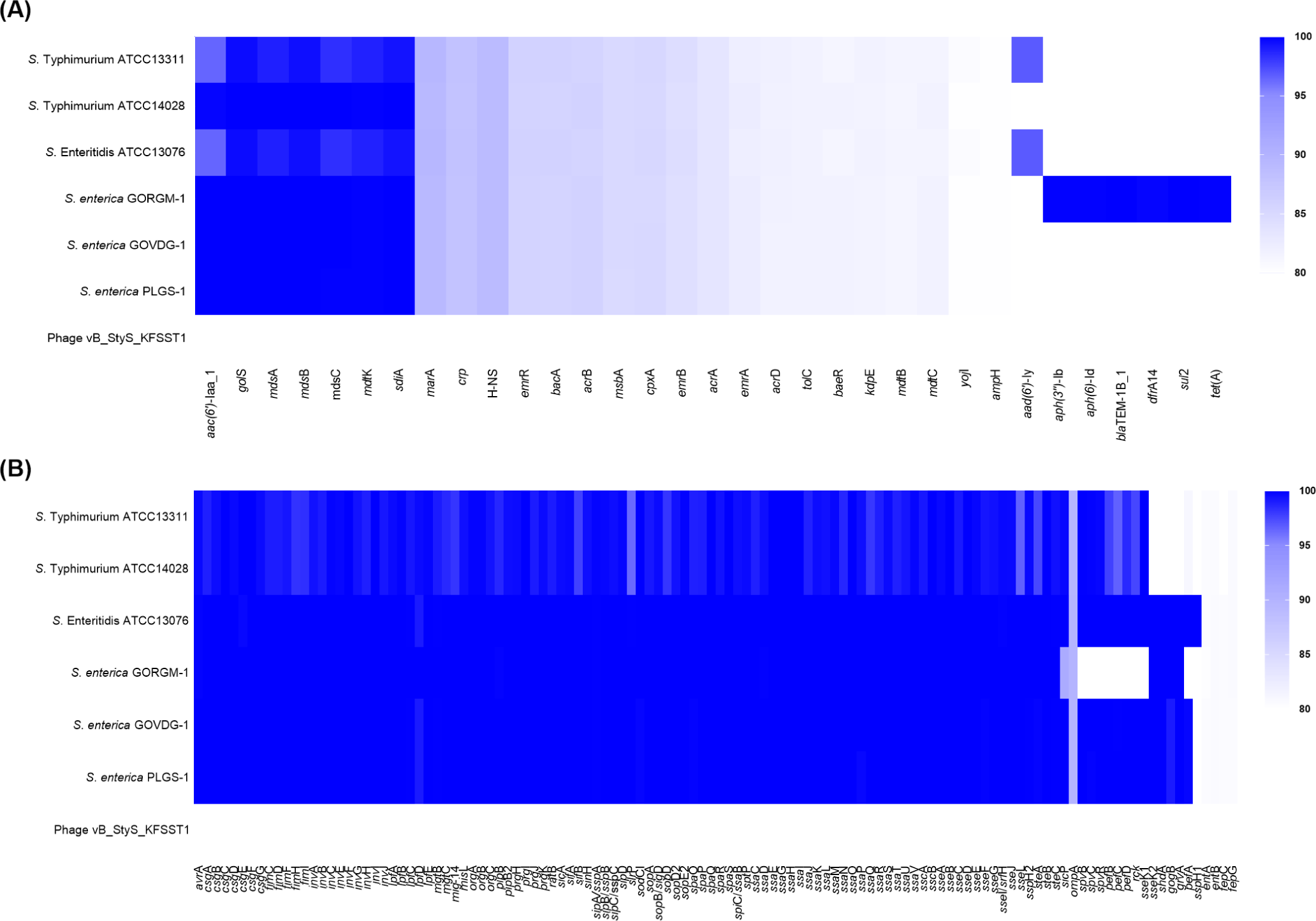
VFDB-based screening revealed that host genomes harbored a wide range of virulence genes (Fig. 3B). Numerous genes related to Salmonella pathogenicity islands, including invA−J, sipA−E, sopA−E, ssaB−U, and prgH−K, were detected in all tested host strain. Other virulence factors such as lpfA−E, sefA, pagC, spvB, spvC, and spvR were also identified in the bacterial genomes. These genes are known to play critical roles in pathogenic mechanisms of Salmonella, including epithelial cell adhesion and invasion (lpfA, sefA, inv, sip, and sop), intracellular survival (ssa and pagC), and systemic infection enhancement (spvB and spvC; Liu et al., 2023; Lou et al., 2019; Marcus et al., 2000). Importantly, no virulence-associated genes could be identified in the genome of vB_StyS_KFSST1.
Although lytic phages are generally considered safer biocontrol agents than temperate or lysogenic phages, recent studies have reported that even lytic phages can occasionally mediate generalized transduction of host DNA fragments, leading to horizontal gene transfer (Fillol-Salom et al., 2018; Schneider, 2021). These findings underscore the necessity of thorough genomic screening when developing phages for biocontrol or feed additive applications. Compared to previous EFSA evaluations of Bafasal®, where genomic safety was primarily confirmed based on the absence of lysogenic genes and manufacturing filtration steps (EFSA Panel on Additives and Products or Substances used in Animal Feed et al., 2021; EFSA Panel on Additives and Products or Substances used in Animal Feed et al., 2024), the present study conducted a more comprehensive genomic characterization by encompassing AMR gene screening, virulence factor profiling, and prophage detection. The complete absence of AMR genes, virulence-associated factors, and prophage-related sequences in the phage genome proposed its excellent genetic stability and minimal biosafety risks. These characteristics align with EFSA guidelines for phage-based feed additives (EFSA Panel on Additives and Products or Substances used in Animal Feed et al., 2024), supporting the potential application of vB_StyS_KFSST1 as a safe and effective candidate for controlling Salmonella in poultry farming. The GenBank accession number of vB_StyS_KFSST1 is PV659140.
The genomic similarity of vB_StyS_KFSST1 to other phages was evaluated based on ANI and phylogenetic analysis. The phylogenetic analysis constructed using genome-BLAST distance phylogeny (GBDP) analysis revealed that vB_StyS_KFSST1 clustered closely together with Salmonella phages KFS-SE2 (GenBank No. NC054641), VSt472 (GenBank No. NC054644), and VB_StyS_B55 (GenBank No. NC054646; Fig. 4). These phages were previously classified within the genus Skatevirus under the family Unclassified Caudoviricetes according to the latest ICTV taxonomy (Simmonds et al., 2024). In contrast, several phages infecting Escherichia coli and other bacterial hosts formed separate clades, confirming the host specificity of vB_StyS_KFSST1.
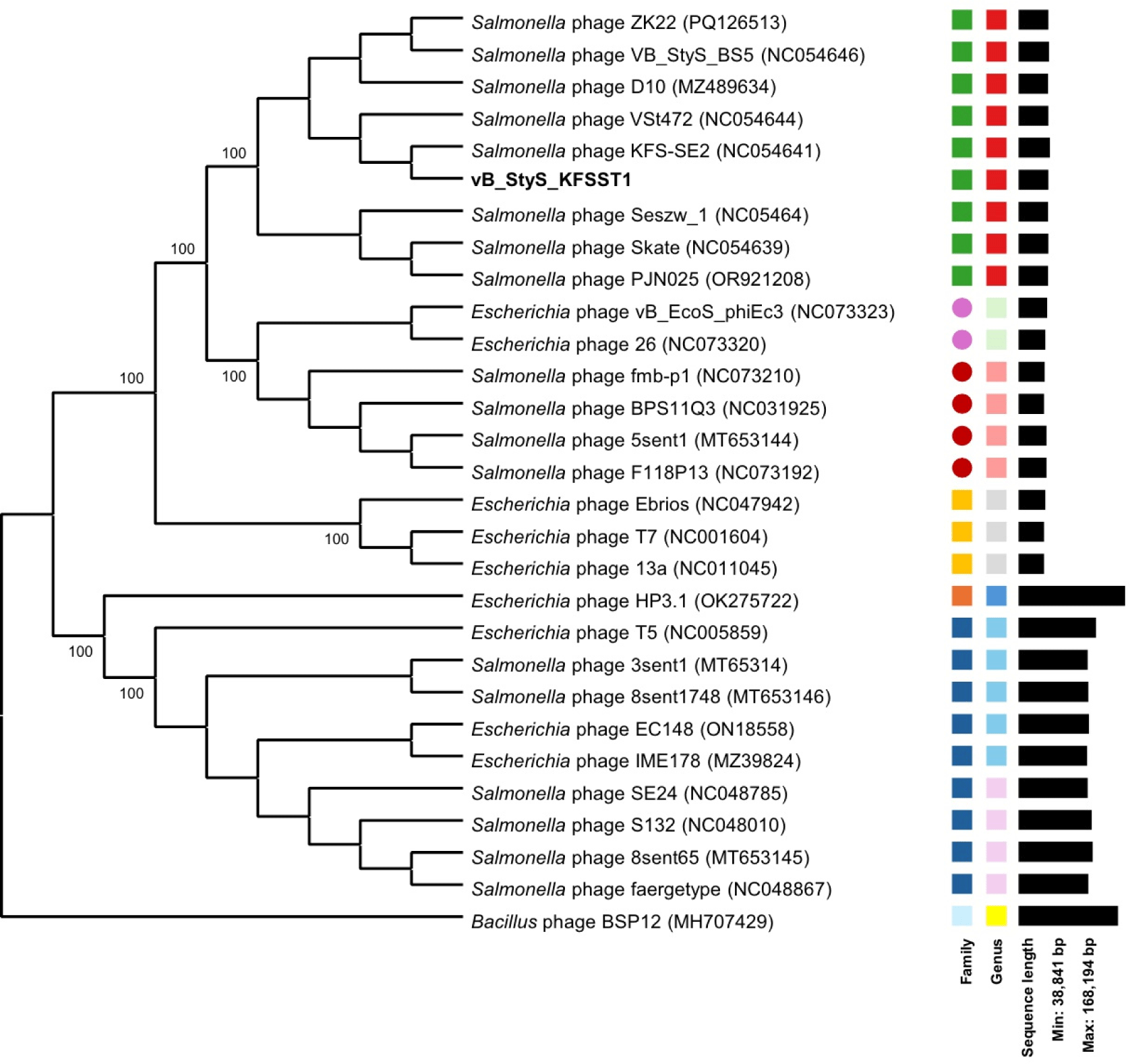
The ANI-based heatmap further supported these findings, showing that vB_StyS_KFSST1 exhibited ANI values over 95% similarity with KFS-SE2, VS47Z, and VB_StyS_B55 (Fig. 5). According to the accepted ANI threshold for species delineation in phages (Adriaenssens and Brister, 2017; Valencia-Toxqui and Ramsey, 2024), these results indicate species-level clustering. Lower ANI values were observed with phages belonging to different genera or different host strain such as E. coli, reinforcing the distinct genomic relatedness of vB_StyS_KFSST1 within the Skatevirus group.
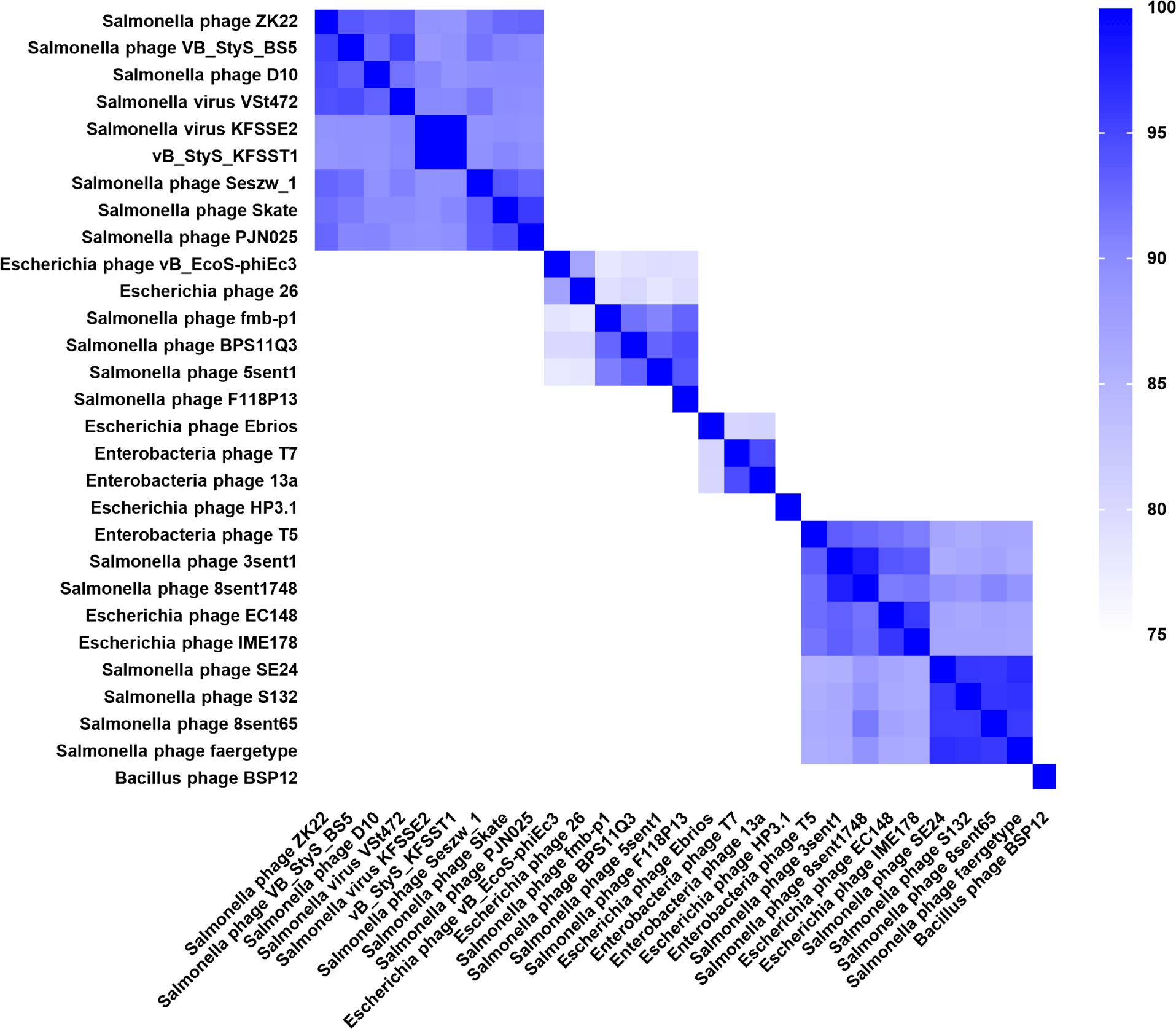
Phylogenetic and taxonomic analyses demonstrated that vB_StyS_KFSST1 belongs to the same species group as Salmonella phages KFS-SE2, VSt472, and VB_StyS_B55 within the genus Skatevirus. Although vB_StyS_KFSST1 is genetically closely related to these Salmonella phages, it exhibits distinct phenotypic characteristics. Among the genetically related phages, KFS-SE2 has been reported to specifically infect S. Enteritidis, showing no lytic activity against S. Typhimurium (Choi et al., 2019). Similarly, PSH-1, a phage closely related to VSt472 with >99% similarity, demonstrated lytic activity primarily against multidrug-resistant S. Enteritidis strains, but did not show any activity against S. Typhimurium strains (Li et al., 2024). Although the phenotypic properties of VB_StyS_B55 were not described, comparative genomic analysis with related phages suggested that the dual serotype-specific activity of vB_StyS_KFSST1 differentiates it from genetically related phages and highlights its potential as a distinct biocontrol candidate.
Conclusion
This study assessed the functional and genomic features of Salmonella phage vB_StyS_KFSST1 to determine its suitability as a candidate for a feed additive in poultry husbandry. The phage exhibited dual serotype-specific and efficient lytic activity against S. Enteritidis and S. Typhimurium, which are two major serotypes associated with poultry-related salmonellosis. Infection kinetics of the phage, marked by rapid adsorption and sustained inhibition of bacterial growth for up to 12 h, confirmed its high in vitro efficacy. Additionally, genome analyses of vB_StyS_KFSST1 confirmed the absence of lysogenic-associated elements, antibiotic resistance genes, and virulence factors, supporting its strict lytic nature and safety. Phylogenetic and ANI-based analyses assigned vB_StyS_KFSST1 to the genus Skatevirus, with distinct phenotypic features compared to closely related phages. These findings support the potential use of vB_StyS_KFSST1 as a safe and effective feed additive candidate for controlling Salmonella in poultry husbandry. Further in vivo validation will be essential to facilitate its practical application and regulatory approval in the livestock industry.













