Introduction
The production of recombinant proteins has been the foundation of the industrial and pharmaceutical biotechnology for the past 30 years (Puetz and Wurm, 2019). There are a wide variety of cell factories, such as mammalian cell lines, insect cells, whole plants, yeast, and bacteria (Ferrer-Miralles and Villaverde, 2013). Lactococcus lactis subsp. lactis IL1403 is a laboratory strain that is widely used in recombinant DNA technology (Pedersen et al., 2005). L. lactis is generally recognized as safe by the Food and Drug Administration (FDA; Song et al., 2017). Moreover, L. lactis is a popular microbial factory system owing to the wealth of genetic knowledge about it, and several existing recombinant protein expression systems (Linares et al., 2010). With the emergence of safety issues related to live microbial cells, including living modified organisms (LMOs), there is increasing interest in probiotic cell components and metabolites (Teame et al., 2020). To date, there have been few studies on the oral administration of bacterial cell extracts as delivery vectors containing recombinant proteins.
Microfold (M) cells are specialized immune cells located in gut-associated lymphoid tissue (GALT), and play an essential role in the initiation of the intestinal immune response that transports luminal antigens through the intestine toward GALT (Foussat et al., 2001). Many previous studies have shown that the receptor activator of NF-kB ligand (RANKL) can induce the differentiation of M cells (Kanaya et al., 2012; Knoop et al., 2009; Kunisawa et al., 2008). Knoop et al. (2009) showed that purified recombinant mouse RANKL produced by a recombinant Escherichia coli differentiated M cells in the small intestine of RANKL-null mice (Knoop et al., 2009). In addition, Kim et al. (2015) showed that oral administration of recombinant L. lactis secreting mouse RANKL significantly increased the number of mature M cells in the mouse small intestine (Kim et al., 2015). These studies demonstrated that recombinant RANKL produced by bacteria is biologically active.
With the advent of next-generation sequencing (NGS) technology, it is easy to determine gene expression levels in the tissues or gut microbiomes. Many studies have focused on the profiling gene expression in diseases, and also profiled the alteration of gut microbiome in diseases or determined the effect of consuming live probiotics, but the alteration of gene expression and gut microbiome after intake of bacterial cell extracts or recombinant proteins is not clear yet.
In this study, the effect of the cell extracts of recombinant L. lactis expressing mRANKL (mRANKL_CE) were evaluated on small intestinal gene expression and gut microbiome in mice, to determine the usefulness of combinations of cell extracts and recombinant proteins for the development of protein drugs.
Materials and Methods
Wild-type L. lactis IL1403 was used as the host strain for recombinant protein production, and grown in M17 medium (MBcell, Seoul, Korea) supplemented with 5 g/L of glucose (M17G). Recombinant L. lactis IL1403 was grown in M17G media with chloramphenicol (5 μg/mL) and erythromycin (5 μg/mL) at 30°C.
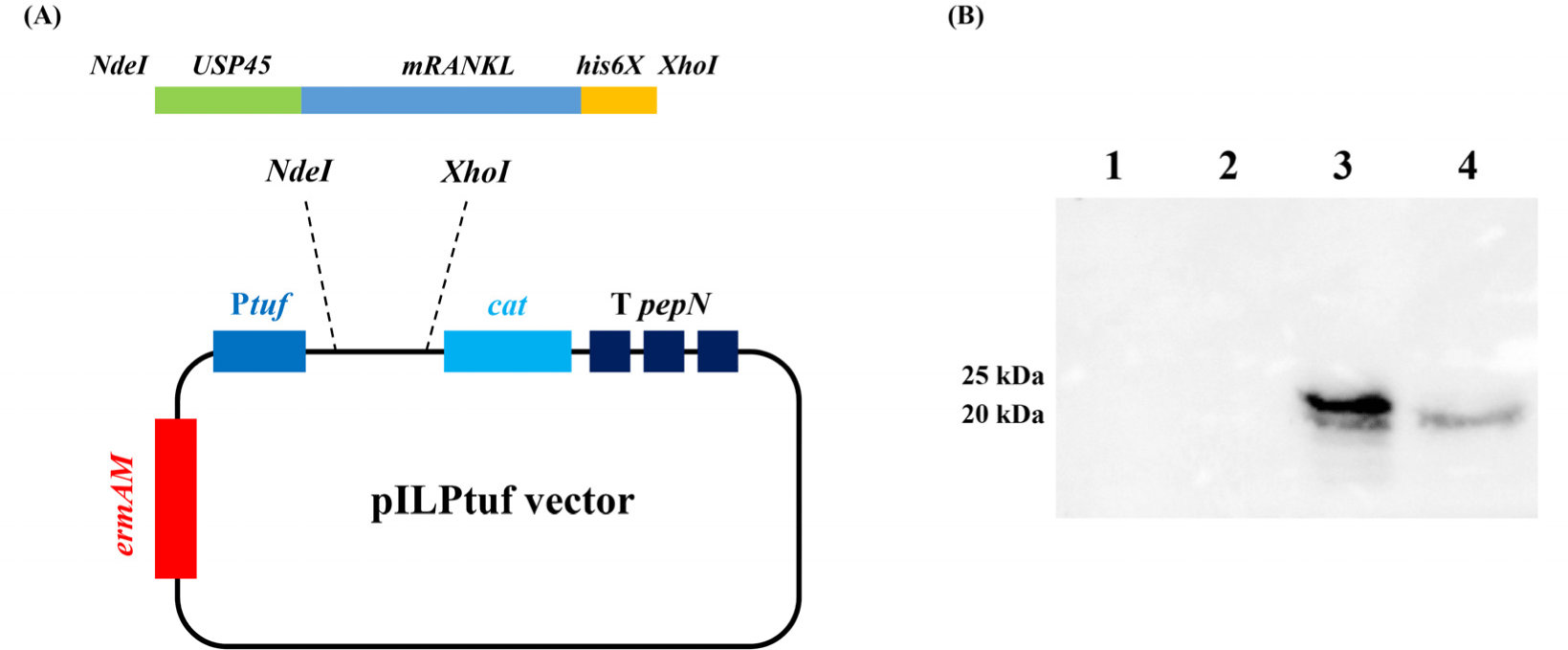
Mouse RANKL sequence was used in Knoop and Kim’s studies (Kim et al., 2015; Knoop et al., 2009). To secrete the target protein, the signal peptide of USP45 (van Asseldonk et al., 1993) was added to the N-terminus, and to detect the expressed protein, a his-tag (his6x) was added to the C-terminus of the target gene. The designed amino acid sequence was codon-optimized using DNAWorks v3.2.4 (Hoover and Lubkowski, 2002) based on the L. lactis IL1403 codon usage table. The insert fragment was synthesized by Macrogen (Seoul, Korea). The insert fragment was shown in Supplementary Fig. S1. The plasmid vector pILPtuf.Mb was used as the backbone (Kim et al., 2009). Insert and vector were ligated at the NdeI and XhoI restriction sites and transformed into wild-type L. lactis IL1403 (WT_CE) competent cells. Vector construction is shown in Fig. 1A.
Wild-type and recombinant L. lactis strains were cultured in 50 mL of M17G broth without and with antibiotics, respectively. The optical density of each sample was measured at a wavelength of 600 nm every 1 h for 12 h and 24 h after inoculation.
The expression of mRANKL from recombinant L. lactis was measured by sodium dodecyl sulphate-polyacrylamide gel electrophoresis (SDS-PAGE) and western blotting. Wild type and recombinant L. lactis were cultured in M17G broth without and with antibiotics at 30°C for 10 h, respectively. For preparation of cell extracts, 10 mL of cultured cells were harvested by centrifugation at 12,300×g for 1 min and lysed using a bead beater with 0.5 g sterilized glass beads (0.5 mm) and 200 μL 1×phosphate buffered saline (PBS). The secreted proteins were collected from 10 mL of cultured cell-free supernatant (0.2 μm filtered) through trichloroacetic acid precipitation, and dissolved in 200 μL 1×PBS for SDS-PAGE and western blot analysis. To quantify the amount of mRANKL produced, 18 kDa commercial recombinant His-tagged human calmodulin (Merck KGaA, Darmstadt, Germany) was used for construction of the standard curve using amounts of 1.5, 1, and 0.5 μg.
RAW 264.7 cells were seeded on cell culture dishes in Dulbecco’s modified Eagle’s medium (DMEM; Capricorn, Düsseldorf, Germany) with 10% fetal bovine serum (FBS; Capricorn) and 1% penicillin and streptomycin (P/S) at a density of 2×106 cells/mL. After seeding for 4 h, the medium was changed to DMEM with 10% FBS, 1% P/S, and 30 ng/mL macrophage colony-stimulating factor with crude cell extracts of wild-type or recombinant L. lactis, that were prepared as described above. The group treated with 1×PBS as control was named PBS, and groups treated with cell extracts from wild type L. lactis IL1403 and mRANKL producing L. lactis were named WT_CE and mRANKL_CE, respectively. According several dose test, finally, the cell extracts containing 90 ng/mL of mRANKL used in this study, and the same amount of cell extracts of WT_CE were used. Commercial mouse RANKL (Abcam, Hanam, Korea) was added to the medium at a concentration of 60 ng/mL.
After 72 h of treatment, the medium was replaced with fresh media, and the cells were incubated for another 72 h. Total RNA was extracted using TRIzol® (ThermoFisher, Seoul, Korea) according to the manufacturer’s instructions, and cDNA was synthesized using PrimeScriptTM RT reagent Kit (Takara Bio, Shiga, Japan). qRT-PCR was conducted using TB Green® Premix Ex TaqTM (Tli RNaseH Plus, Takara Bio) with specific primers TRAP-F:5′-GCGACCATTGTTAGCCACATACGG-3′; TRAP-R:5′-CGCCCAGGGAGTCCTCAGATCCAT-3′ and primers for the housekeeping gene GAPDH-F:5′-AACTTT GGCATTGTGGAAGGGCTC-3′; GAPDH-R:5′-AAGGCCATGCCAGTGAGCTTC-3′. mRNA levels were normalized to those of GAPDH (Kim et al., 2015).
Four-week-old BALB/c mice were randomly assigned to the three groups (PBS, WT_CE, and mRANKL_CE). Before oral administration, 300 μL of neutralizing reagent (1.5% NaH2CO3) was administered to prevent gastric acidity. After 30 min, 100 μL 1×PBS was fed to the PBS group, and the cell extracts from 2.5×108 CFU wild type L. lactis IL1403 and cell extracts containing mRANKL (4.2 μg) from 2.5×108 CFU of recombinant L. lactis were fed to WT_CE and mRANKL_CE groups, respectively. All groups were fed for seven consecutive days and sampled on the eighth day (Supplementary Fig. S2).
Peyer’s patches of small intestine samples were extracted to determine the abundance of mature M cells by measuring the GP2 mRNA expression level. Isolation of RNA and synthesis of cDNA was same as above method. qRT-PCR was conducted using TB Green® Premix Ex TaqTM (Tli RNaseH Plus, Takara Bio) with M cell-specific primers GP2-F:5′- GATACTGCACAGACCCCTCCA-3′; GP2-R:5′- GCAGTTCCGGTCATTGAGGTA-3′ (Kusunose et al., 2020), and primers for the housekeeping gene GAPDH-F:5′-AACTTTGGCATTGTGGAAGGGCTC-3′; GAPDH-R:5′-AAGGCCATGCCAGTG AGCTTC-3′. mRNA levels were normalized to those of GAPDH (Kim et al., 2015).
Nine ileum samples of the same size (1 cm) were extracted from the same position (distal ileum). Total RNA was isolated from the tissues using the Maxwell (Promega) method. One milligram of total RNA was processed to prepare the mRNA sequencing library using the MGIEasy RNA Directional Library Prep Kit (MGI), and sequencing was performed using the MGIseq system.
After a quality check, raw reads were trimmed to 100 bp prior to mapping to the mouse reference genome GRCm38/mm10 using HiSat2 (version 2.1.0; Pertea et al., 2016). Following read alignment, counts assigned to features were computed using the featureCounts (version 2.0.3; Liao et al., 2014). Differentially expressed genes (DEGs) were identified using the DESeq2 (v1.24.0; Love et al., 2014). A gene was considered to be a DEG with a fold change>1.45 and adjusted p-value<0.1 using the Benjamini-Hochberg method (Benjamini and Hochberg, 1995). Gene Ontology (GO) enrichment analysis was performed using in-house Perl scripts. The significantly enriched GO terms were determined by Fisher’s exact test with p<0.05 and odds ratio>1.
Genomic DNA was extracted from fecal samples using a NucleoSpin Soil kit (Macherey-Nagel, Düren, Germany) according to the manufacturer’s instructions. DNA samples (5 ng) were used to amplify the 16S ribosomal RNA V4 region using Takara Ex-taq DNA polymerase (Takara Bio) with universal primer sets (Forward:5′-GGACTACHVGGGTWTCTAAT-3′ and R:5′-GTGCCAGCMGCCGCGGTAA-3′; Han et al., 2018). After amplification, all the samples were normalized to 50 ng per sample. A DNA library was then constructed and sequenced using the Illumina MiSeq platform (Illumina, San Diego, CA, USA), generating 2×300 bp paired-end reads.
To analyze the gut microbiome, de-multiplexed and pre-processed sequence reads were imported into Quantitative Insights Into Microbial Ecology (QIIME2, version 2021.2; Bolyen et al., 2019). Barcode and primer removal, quality control, amplicon sequence data correction, and de-replication, were performed using the DADA2 (Callahan et al., 2016). Feature tables and representative sequence files were merged for downstream analysis using QIIME2. Taxonomic classification was performed using the SILVA 132 database with 99% identity, based on the V4 16S region. All classification was performed within QIIME2 and was assigned using the naïve Bayesian algorithm available in the sklearn python library. For phylogenetic diversity analysis, alpha and beta diversities were calculated using the q2-diversity plugin, and included Faith’s phylogenetic diversity and weighted and unweighted UniFrac distances. Differential abundance analysis of microbiota was performed using an in-house Perl script. We considered a p-value<0.05 to indicate statistical significance.
Results
To confirm the expression of mRANKL, intracellular and secreted proteins were precipitated and analyzed by western blotting. No mRANKL was detected in the wild-type L. lactis samples. In recombinant L. lactis, intracellular and secreted mRANKL proteins were detected with expected size of 23.86 and 20.88 kDa, respectively (Fig. 1B). According to the standard curve, recombinant L. lactis produced 2.1 μg/mL of mRANKL in intracellular fraction (Supplementary Fig. S3).
To examine the physiological characteristics of wild type and the recombinant L. lactis, patterns of growth were measured. As shown in Supplementary Fig. S4, recombinant L. lactis showed a slightly delayed growth rate compared to the wild type; after 10 h though, these two strains showed similar growth patterns.
To validate the biological activity of mRANKL, RAW 264.7 cells were used to observe the stimulation of RANK-RANKL signaling. As shown in Supplementary Fig. S5, treatment with culture media containing commercial RANKL and mRANKL_CE significantly (p<0.05) increased TRAP mRNA expression levels compared with those in the PBS and WT_CE groups.
Cell extracts containing recombinant mRANKL were orally administered to mice for seven consecutive days, and the expression of mature M cells marker in Peyer’s patches was analyzed by qRT-PCR. As shown in Supplementary Fig. S6, the GP2 mRNA expression levels in the mRANKL_CE group were significantly (p<0.05) higher than those in the PBS and WT_CE groups. In addition, body weight gain was measured between day 1 and day 8 of the experiment, and the results showed that oral administration of WT_CE and mRANKL_CE had no significant effect on body weight (Supplementary Table S1). Our results showed that recombinant mRANKL from recombinant L. lactis is biologically active and does not cause significant changes in body weight.
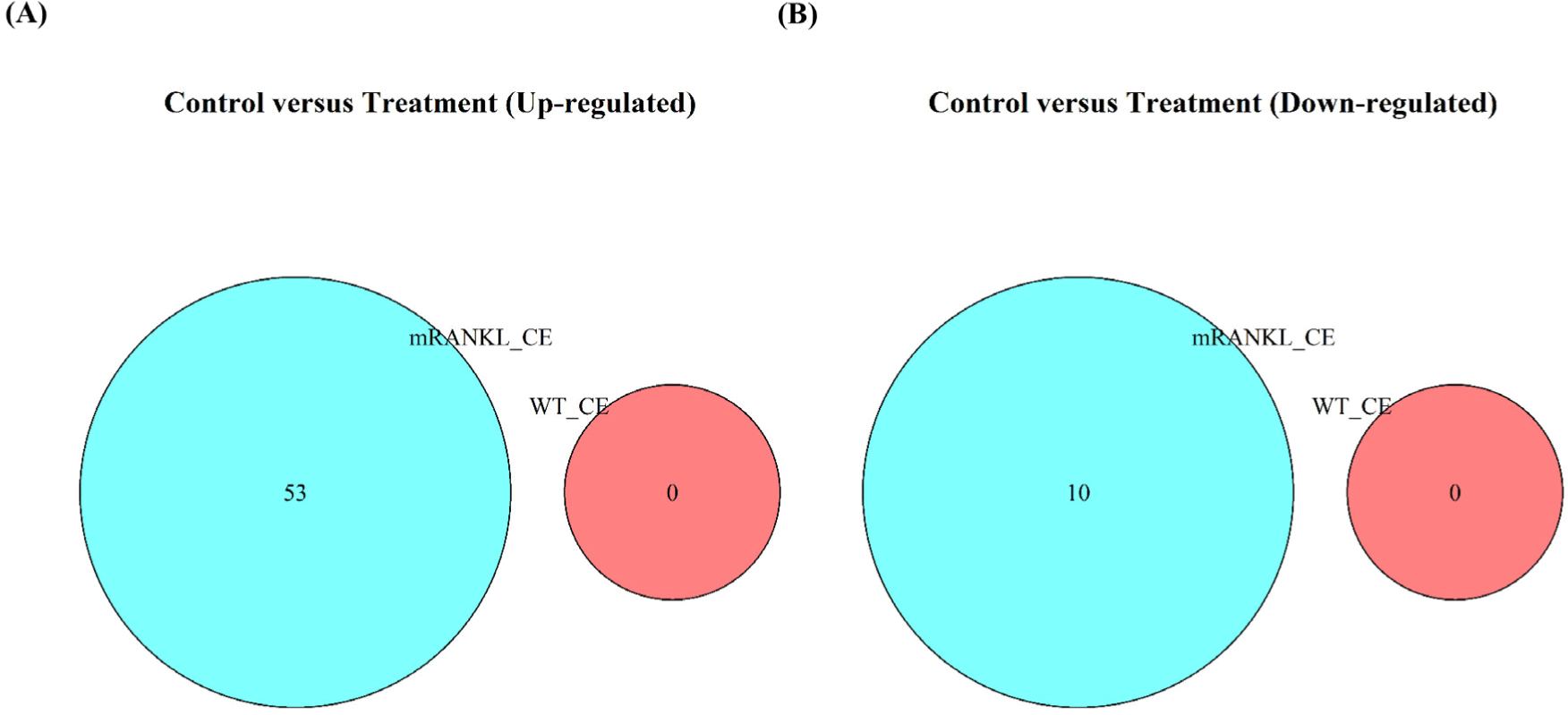
To compare the gene expression between the PBS, WT_CE, and mRANKL_CE groups, RNA-Seq analysis of the mouse small intestine was performed. The WT_CE and mRANKL_CE groups were compared with the PBS group, with a cut-off fold change>1.45 and adjusted p-value<0.1. No DEGs were identified between the PBS and WT_CE groups. Between the PBS and mRANKL_CE groups, 63 DEGs, including 53 upregulated and 10 downregulated DEGs, were identified (Fig. 2 and Supplementary Table S2). Between the WT_CE and mRANKL_CE groups, 192 DEGs were identified, including 169 upregulated and 23 downregulated DEGs (Supplementary Table S2). These results indicate that cell extracts from WT_CE had no effect on mouse small intestinal gene expression, and only mRANKL had an effect.
To analyze the gene enrichment of DEGs, GO analysis were performed with a threshold of p<0.05 and odds ratio>1. Between the PBS and mRANKL_CE groups, 49 upregulated and 9 downregulated DEGs were annotated with GO terms. The upregulated GO terms included ‘de novo’ protein folding (p<0.001), response to unfolded protein (p<0.001), regulation of bone resorption (p<0.01), heat shock protein binding (p<0.001), and calcium ion binding (p<0.05; Table 1 and Supplementary Table S3). Between the WT_CE and mRANKL_CE groups, 159 upregulated and 22 downregulated DEGs were annotated with GO terms. The upregulated GO terms included calcium ion binding (p<0.001), calmodulin binding (p<0.001), and ‘de novo’ protein folding (p<0.05; Table 2 and Supplementary Table S3).
To compare the gut microbial diversity, the alpha and beta diversities of the three groups from normalized microbiome sequencing reads were investigated. For alpha diversity, three indices including observed features, Shannon and Faith’s phylogenetic diversity (Faith PD) were measured. None of the three indices showed any significant differences among the three groups (Supplementary Fig. S7). For beta diversity, principal coordinate analysis (PCoA) of unweighted and weighted UniFrac distances was performed. There were no significant differences among the three groups (Supplementary Fig. S8).
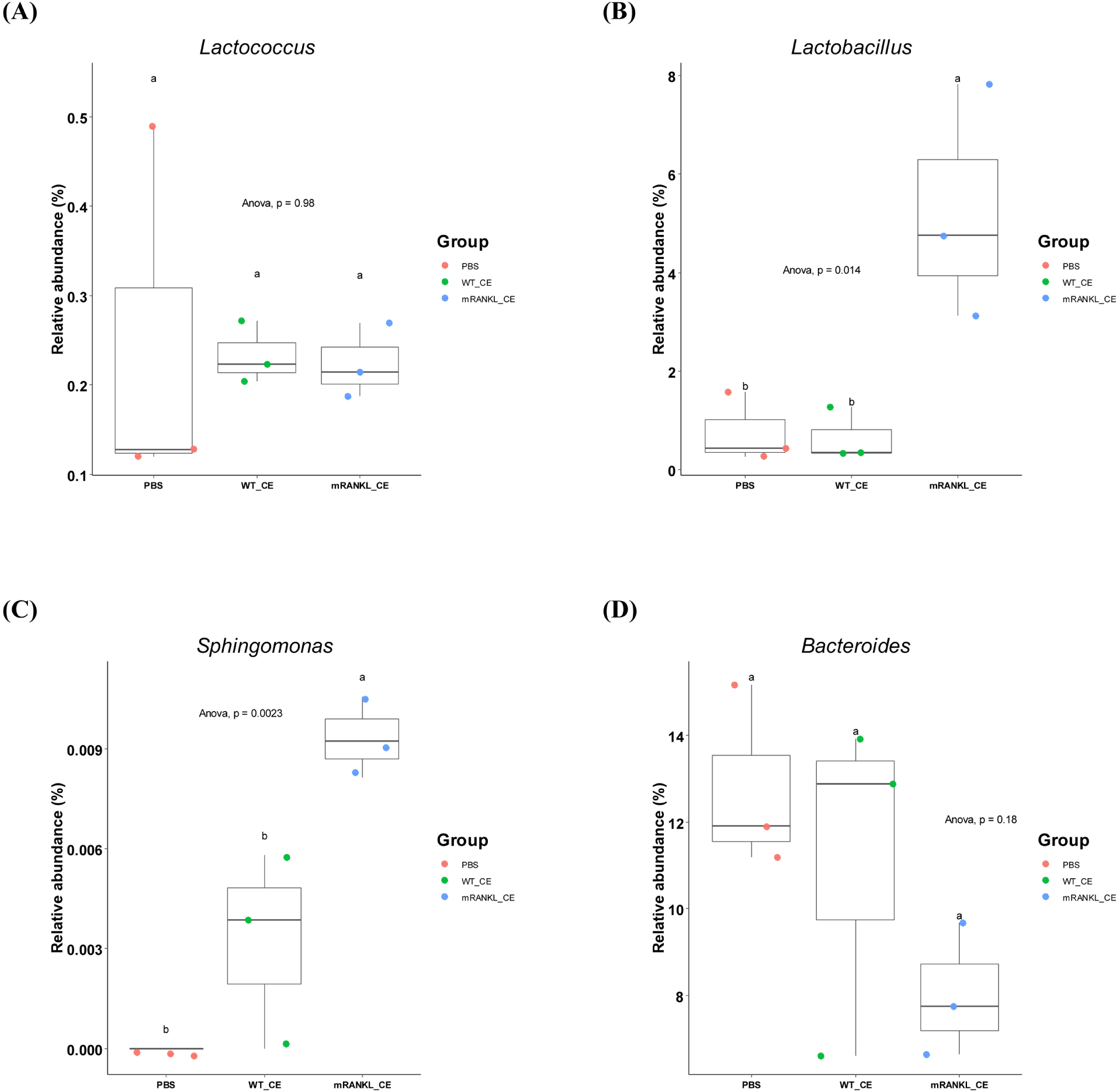
To compare the differences in major gut microbial taxa among the three groups, the microbial composition in these three groups were examined. The overall microbial composition in the gut was not significantly different among the three groups. However, the genera Lactobacillus (p<0.05), Sphingomonas (p<0.01) and Acinetobacter (p<0.01) differed significantly (Fig. 3 and Supplementary Table S4). Moreover, there was no significant difference among the three groups in the genus Lactococcus (Fig. 3 and Supplementary Table S4). These results showed that feeding cell extracts with or without mRANKL did not have a significant effect on the gut microbiome.
Discussion
Many recombinant microorganisms are widely used in food, chemical, and pharmaceutical industries today (Khan et al., 2016). The direct use of LMOs poses potential risks to the environment, humans, and animals (Prakash et al., 2011).
LAB are traditionally used in fermentation and food preservation, and are recognized as safe for consumption. In particular, L. lactis has been engineered as a live vehicle for the delivery of DNA vaccines and production of therapeutic biomolecules (Tavares et al., 2020). In bacterial cell extracts study, cell extracts from recombinant L. lactis expressing SARS-CoV-2 spike protein were used to oral immunize mice, and antigen-specific antibodies were produced from immunized mice (Xuan et al., 2022). L. lactis IL1403 is a representative laboratory strain that can be used to produce recombinant heterologous proteins (Tavares et al., 2020), and there is no evidence to date that metabolites of L. lactis IL1403 are toxic to experimental animals. Moreover, use of cell-free extracts removes the risk of LMOs being released into the environment. Our results demonstrated that cell extracts containing mRANKL from recombinant L. lactis IL1403 can differentiate RAW 264.7 cells into osteoclast-like cells (Supplementary Fig. S5) and increase the number of mature M cells in mice (Supplementary Fig. S6). In addition, in the transcriptome analysis, no DEGs were found between PBS and WT_CE groups; while DEGs were found between PBS and mRANKL_CE groups. These results indicate that WT_CE has no effect on mouse small intestinal transcriptome, whereas mRANKL does have an effect. Moreover, RANKL not only affects the differentiation of M cells, but also affects other RANKL signal-responsive cells (Kukita and Kukita, 2013). Knoop et al. (2009) demonstrated not only GALT expressed RANK in mice (Knoop et al., 2009). Heat shock proteins are a family of proteins produced by cells in response to stress, and they positively regulate osteoclastic bone resorption through the RANKL-RANK signaling pathway (Hang et al., 2018). In this study, genes associated with heat shock protein binding and regulation of bone resorption were found to be upregulated in the mRANKL_CE group. Calcium signaling plays a significant role in osteoclastogenesis; the RANKL receptor utilizes calcium signaling to drive osteoclast differentiation (Komarova et al., 2003), and genes associated with calcium ion binding were found to be upregulated in the mRANKL_CE group. Additionally, SLIT3 was found to be co-upregulated in the mRANKL_CE group compared to its expression in the PBS (adjusted p<0.1) and WT_CE (adjusted p<0.1) groups (Supplementary Table S2). SLIT3 expression increases during osteoclast differentiation (Koh, 2018). In addition, bone remodeling-associated genes ZBTB16 (vs. PBS, adjusted p<0.001; vs. WT_CE, adjusted p<0.01) and ZBTB40 (vs. PBS, adjusted p<0.01; vs. WT_CE, adjusted p<0.1) were found to be co-upregulated in the mRANKL_CE group (Supplementary Table S2; Felthaus et al., 2014; Twine et al., 2016). However, no study so far has investigated whether an increase in RANKL levels in the intestine causes bone resorption. Since the cell extracts of the host producing recombinant proteins do not appear to have any effect on experimental animals, this strategy may be an alternative to the use of live cells. The well-known signal peptide USP45 is located at the upstream of the target protein and its secretion is relatively low. Improving the secretion yield of target proteins in cell-free culture supernatant, can be a good alternative to the direct use of LMOs.
Recently, some studies have used postbiotics replace the live bacteria to improve the intestinal environment; postbiotics are defined as “non-viable bacterial products or metabolic products from microorganisms that have biological activity in the host” (Nataraj et al., 2020; Siciliano et al., 2021). Postbiotics contain a wide range of molecules, including peptidoglycans, surface proteins, cell wall polysaccharides, secreted proteins, bacteriocins, and organic acids, which can have positive effects on the host, including immunomodulatory, antitumor, antimicrobial, and barrier preservation effects (Nataraj et al., 2020; Teame et al., 2020). In our study, bacterial crude cell extracts had no adverse effect on the host, but exogenous proteins had favorable effects. Hence, exogenous protein can be produced by probiotics, and the cell extract can then be used for therapeutic or other purposes. Also, the crude cell extracts can be used directly, without the need for purification (Taghinezhad-S et al., 2021).
There have been many recent studies on the host-microbiome interactions in postbiotics (Nataraj et al., 2020; Peluzio et al., 2021; Siciliano et al., 2021; Teame et al., 2020). However, there have been no studies on the state of intestinal microbiome after intake of live recombinant LAB or its cell extracts. In this study, NGS were used to characterize how cell extracts of wild-type or recombinant L. lactis affected the gut microbiome. Although there was no significant change in the gut microbiome, the abundances of several genera were significantly different (Fig. 3 and Supplementary Table S4). RANKL stimulates the differentiation of monocyte / macrophage precursor cells into osteoclasts, and overexpression of RANKL resulting in bone erosion in rheumatoid arthritis (RA; Tanaka, 2019). The abundance of Lactobacillus is significantly higher in patients with RA than in healthy controls (Li et al., 2021). Our results also showed that the abundance of Lactobacillus was significantly (p<0.05) higher in the mRANKL_CE group. The abundance of Sphingomonas was also significantly (p<0.01) higher in the mRANKL_CE group. Eriksson et al. (2022) showed that Sphingomonas abundance was positively correlated with RA (Eriksson et al., 2022). Another RA-related genus, Bacteroides, showed decreased abundance in the mRANKL_CE group, although the difference was not significant. In addition, the abundance of Bacteroides was decreased in patients with RA compared with its abundance in the healthy controls (Wang et al., 2022). These results may indicate changes in abundances of specific microbes caused by excessive RANKL in the intestine. One diagnostic marker in RA patients is a higher serum calcium concentration than that in the healthy controls (Tawfik et al., 2019). In addition, an increase in RANKL levels in the bone microenvironment leads to bone resorption and increased calcium release (Ono et al., 2020). Our result indicated no significant differences in serum calcium levels among the three groups (Supplementary Table S5). Moreover, the abundance of Lactococcus, which was used as the host for the production of mRANKL, was not significantly different among the three groups (Fig. 3). This indicates that the cell extracts of L. lactis did not elicit an immune response, which may be because Lactococcus is a natural inhabitant of the mouse gut. Our results indicate that the cell extracts of L. lactis did not have a significant impact on the mouse gut microbiome.
Conclusion
In summary, the cell extracts containing mRANKL from recombinant L. lactis are biologically active both in vitro and in vivo, and the cell extracts from WT_CE did not affect intestinal transcriptome. In addition, there was no significant change in the gut microbiome after administration of cell extracts of WT_CE or recombinant L. lactis. This strategy could potentially be used for the development of protein drugs.
Supplementary Materials
Supplementary materials are only available online from: https://doi.org/10.5851/kosfa.2022.e54.













