Introduction
Due to improvements in economic drivers for local communities, demand for animal products and particularly meat, are increasing in order to meet dietary demands. Especially, chicken meat is relatively cheap and a good source of animal protein for human health. However, as more and more chicken meat is being consumed, there are concerns about food safety and security. Chicken meat has a short shelf life because psychrotrophic bacteria causes spoilage or off-flavors even at cold storage conditions (Carrizosa et al., 2017). The spoilage of meat depends on pH level, availability of oxygen, biodiversity of bacterial groups, and storage temperature (Ercolini et al., 2009). These factors, in turn, are closely associated with the growth of spoilage bacteria.
In food industries, the increase in contamination of meat is a major issue at slaughter and during processing (Remenant et al., 2015). In addition, increased pH after rigor mortis induces bacterial growth of putrefactive bacteria. Pseudomonas spp. is a major psychrotrophic bacteria that produces proteinase and its optimal pH is from 6.5 to 8.0. Proteinase hydrolyses chicken protein and causes spoilage (Fairbairn and Law, 1986; Nowak et al., 2012). Availability of oxygen generally affects the microbial populations that will proliferate. When oxygen levels are high, aerobic or facultative anaerobic gram-negative bacteria dominate but at low oxygen levels, facultative anaerobic or anaerobic gram-positive microbiota grow (Doulgeraki et al., 2012).
Storage temperature, however, is the most important factor that affects the growth of bacteria present in chicken meat. Psychrotrophic bacteria can grow at refrigerated conditions, and temperature can affect various microbial growth parameters including maximum growth rate and total bacterial counts (Mataragas et al., 2006). Doulgeraki et al (2010) reported that storage temperature can affect the spoilage potential of bacteria and that different strains of the same species do not necessarily grow at the same rates.
Traditionally, microbial populations of meat has been analyzed by culture-dependent plating methods. It is based on the isolation and culturing of bacteria from chicken meat and identification by 16S rDNA sequencing. This method has limitations in that not all bacteria present in chicken meat can be cultivable on the plate media used in the microbial analysis (Grama et al., 2002). However, in recent studies, molecular technologies are being developed with much higher resolving power to advance research (Doulgeraki et al., 2012). Next generation sequencing (NGS) could play an important role in understanding the in-depth biodiversity of spoilage microbiota by analyzing many different samples. Also, NGS can discriminate taxa to specific strains allowing for detailed analysis of bacteria present in meat (Broekaert et al., 2011; Casaburi et al., 2011; Diez et al., 2008).
The objectives of this study were to characterize the putrefactive bacteria isolated from chicken meat during cold storage and to analyze the composition of chicken meat spoilage microbiota using NGS.
Material and Methods
Chicken meat was purchased commercially as whole chicken (with skin) and chicken breast in local markets. To test for spoilage, two types of chicken meat were refrigerated at 4°C and 10°C. The chicken samples were kept for 5 to 15 d until spoilage. After storage, phosphate buffer saline (PBS) at 2 X the sample weight was added to samples and was homogenized using a stomacher (3M, Korea) at maximum speed for 2 min. The supernatant was then filtered through sterilized gauze to remove floating matter.
The filtrate was serially diluted 10-fold with PBS solution, and one hundred microliters was inoculated onto Tryptic Soy Agar (TSA, BD Difco, USA). Inoculated plates were incubated at 25°C for 24 to 48 h. After incubation, colonies were randomly selected from the most diluted plates for the isolation of psychrotrophic bacteria. The isolates were cultured and stored at −80°C in 10% skim milk media.
For this analysis, the isolates were amplified using the PCR process and isolates were identified by 16S rDNA sequencing (Solgent Co., Korea). We used universal primers 27F and 1492R with 2x H-star Taq PCR master mix (Biofact Co., Ltd, Korea). The sequences were confirmed using the BLAST search and EzBioCloud (Yoon et al., 2017).
For the analysis of bacterial communities, bacterial DNA was extracted from the chicken meat filtrates. The filtrates were centrifuged at 13,000 rpm for 10 min and re-suspended with PBS. Bacterial DNA was extracted using QIAamp powerfecal DNA isolation kit (Qiagen, Netherlands). DNA concentration and purity was evaluated for subsequent analysis and samples were sequenced by Illumina MiSeq platform and identified using CLcommunity software (Chunlab, Korea).
Isolated psychrotrophic bacteria were measured for growth at different temperature and by media. The isolates were inoculated on TSA and Plate Count Agar (PCA, BD Difco, USA) and incubated at 10°C, 25°C, and 37°C. The plates were stored for 7 d and bacterial growth was measured at 24 h intervals.
Skim milk agar (SMA, 10% of skim milk solution and 1.5% of Bacto agar [BD Difco, USA]) and chicken juice agar (CJA, 50 g chicken breast meat ground in 250 mL sterilized deionized-water and filtered through two layers of gauzes; the filtrate were kept in a water bath at 50°C) was used to compare proteolytic activity of the isolates. For making agar plates, 250 mL of 3% Bacto agar solutions were prepared at 50°C and mixed it with the filtrate solution prepared as above (Wang et al., 2017). The isolated psychrotrophic bacteria were inoculated on SMA and CJA plates, and incubated at 25°C for 2 d. After incubation, the plates were examined for clear zones around colonies.
Results and Discussion
For the isolation of psychrotrophic bacteria, 12 samples of chicken meat were stored at 4°C and 10°C, and the period of storage was continued until observing similar patterns of spoilage on the surface including microbiological, visible and organoleptic features. During the experimental conditions, the number of bacteria increased gradually with storage time. After 5 d, bacterial growth in chicken meat increased over 8.3 Log CFU/g. In addition, it was confirmed that significantly more bacterial growth was observed on whole chicken than on chicken breast. According to Carrizosa et al. (2017), putrefactive bacteria at concentrations more than 8 Log CFU/g can cause spoilage and off-flavor due to the degradation of amino acids.
For the isolation of psychrotrophic bacteria, colonies were randomly selected from TSA plate at the most diluted concentrations. We isolated 118 strains from chicken meat and they were incubated at 25°C for > 12 h before use. The identification of bacterial isolates was carried out using 16S rDNA sequencing. Because bacteria have conserved regions in common, they can be identified after PCR amplification based on universal primers (Nocker et al., 2007).
From the culture-dependent approach, we isolated 118 strains of psychrotrophic bacteria and they comprised Pseudomonas spp. (58.48%), Serratia spp. (10.17%), and Morganella spp. (6.78%) (Fig. 1). In total, 16 different species of Pseudomonas were identified with Pseudomonas weihenstephanensis and Pseudomonas psychrophila the most abundant (Table 1). Pseudomonas weihenstephanensis has been recently reported as a novel species from raw cow's milk (von Neubeck et al., 2016). It is similar to Pseudomonas fragi and P. psychrophila based on phylogenetic analyses, and has a potential to cause spoilage of food. In general, Pseudomonas spp. mainly appeared at low temperatures and aerobic conditions, and species of this genus such as P. fragi, P. lundensis, and P. fluorescens have been reported in the putrefaction of meat (Casaburi et al., 2015; Lebert et al., 1998).
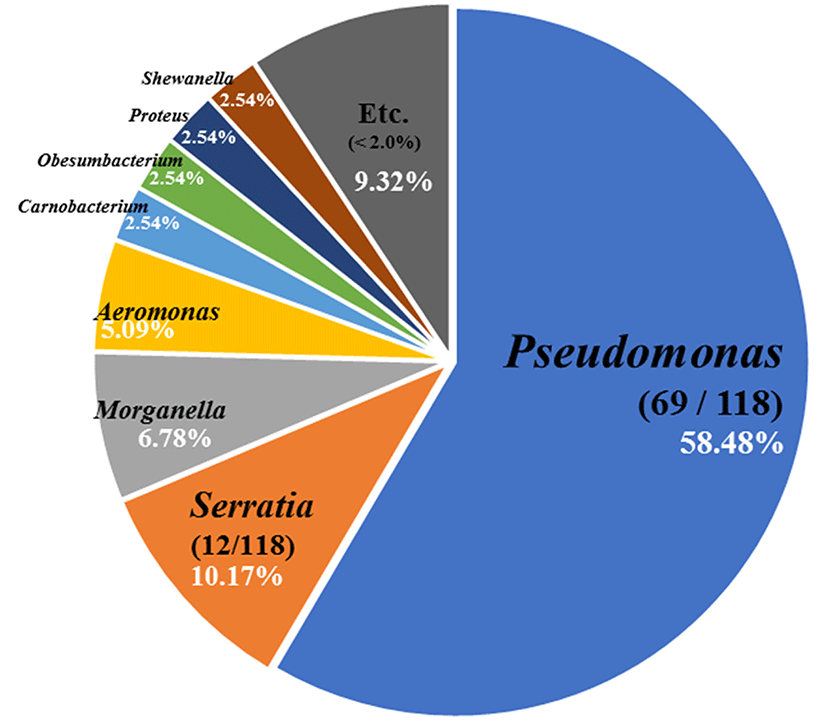
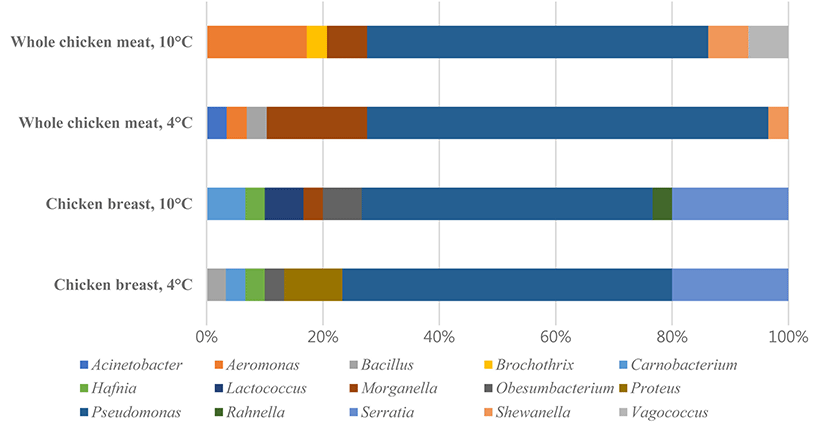
All experiments were divided into four treatment groups, which were composed of two different types of chicken meat (chicken breast, CB; whole chicken meat, CS) and two different temperatures (4°C and 10°C). The most frequently observed bacteria in the microbial populations of each treatment group were Pseudomonas spp., and Serratia spp., Carnobacterium spp., Hafnia spp., and Obesumbacterium spp. Especially, Serratia liquefaciens and S. proteamaculans were the most predominant species in air packed meat (Pennacchia et al., 2011).
In addition, the treatment groups were compared to evaluate differences in bacterial composition. Fig. 2 shows the composition of isolated psychrotrophic bacteria and indicates that CB4 and CB10 exhibited similar patterns than other treatments. Also, bacterial sharedness was measured which is an index that represents different groups sharing common microbiota. It was demonstrated that CB 10-CB4 had the highest score (80%), followed by CS10-CS4 with 72% and implies that the psychrotrophic bacteria was more affected by different meat than the storage temperature.
To identify bacterial communities, bacterial DNA was extracted from each treatment group and analyzed by an Illumina MiSeq platform and 187,671 reads were obtained. Pseudomonas spp. comprised ~85% followed by Serratia spp. (5.95%), Acinetobacter spp. (3.45%), and Morganella spp. (1.06%) (Fig. 3). In addition, P. weihenstephanensis (34.59%) and P. congelans (13.74%) were the most prominent species in the NGS analysis (Table 2).
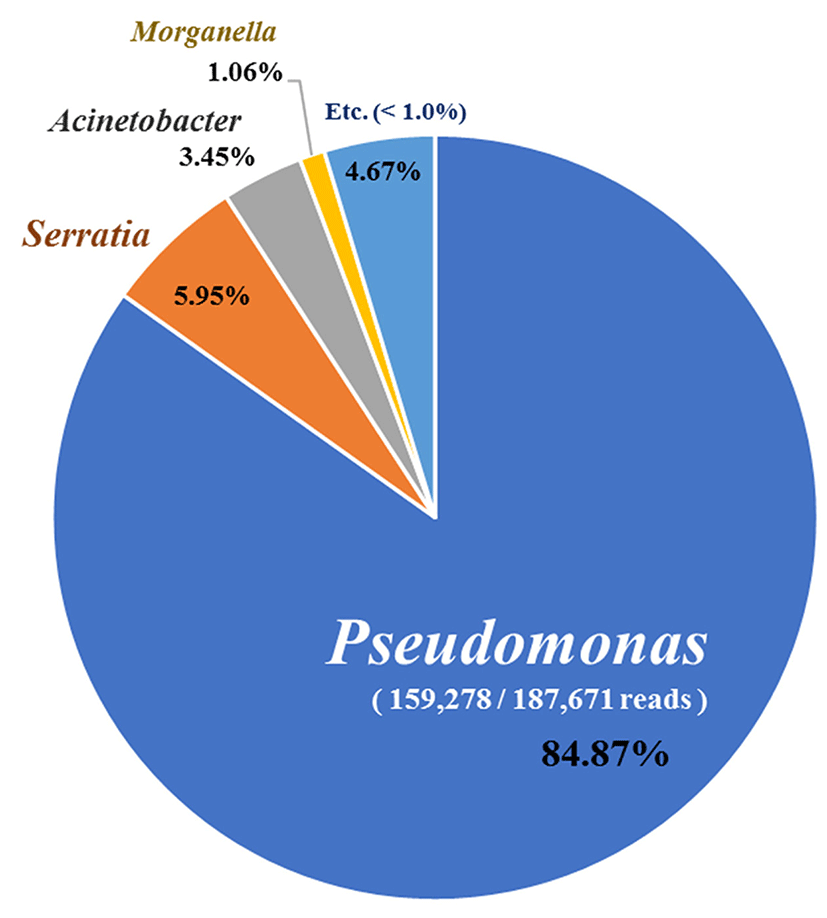
Although the biodiversity of this bacterial community was not highly correlated to the psychrotrophic bacteria, they shared similar patterns. Predominant genera such as Pseudomonas, Serratia, and Morganella were identified from both experiments. Among these two analyses, the advantage of using NGS was the fact it had higher resolving power than the culture-dependent approach. For example, traditional methods of analyzing microbial population are species-specific species but NGS technology can evaluate bacterial communities from environmental samples (Wang et al., 2017). In our study, the culture-dependent approach identified only 16 species of Pseudomonas whereas NGS provided 111 species. Consequently, advanced metagenomic technology could play an important role in understanding the biodiversity of spoilage microbiota in chicken meat (Doulgeraki et al., 2012).
To evaluate differences in bacterial communities, MiSeq sequences were analyzed by CLcommunity programs and their relationships were evaluated using sharedness and depicted by UPGMA dendrograms. The bacterial similarity between CB10 and CB4 was much closer than the other populations. The sharedness of each treatment was 80.13% between CB10-CB4 and 80.61% between CS10-CS4. The amount of sharedness was, however, higher than CB4-CS4 (74%) and CB10-CS10 (67%) at the same storage temperature. This indicates that the type of chicken meat was a greater influence than the storage temperatures on bacterial communities. It was assumed that whole chicken (with skin) could be contaminated from feathers, feces, and soil, so that it might harbor different microbial populations than chicken breast (Russell, 2009; Tuncer and Sireli, 2008).
The isolated psychrotrophic bacteria from chicken meat was composed of many species, and strains of the same species. The isolates were assessed for the growth at different temperatures and media, and also evaluated for proteolytic activity to predict their putrefactive potential.
Almost all of the isolated psychrotrophic bacteria were able to grow at 10°C and 25°C but some of these bacteria did not grow at 37°C even after 5 d. This is probably due to their psychrophilic characteristics.
For the assessment of proteolytic activity, activity was identified by a clear zone around the colonies (Fig. 4). In this experiment, some species were shown to exhibit similar proteolytic activities in both media. However, Proteus spp. only able to cause the clear zone in skim milk agar. About two-thirds of Pseudomonas spp. showed some degree of proteolytic activity, and the highest activity was exhibited by Serratia spp. and Aeromonas spp. that were able to hydrolyze proteins in the plates. However, Morganella spp. of all isolates showed no activity (Table 3). These strains were considered non-spoilage bacteria although it is apparent they can grow at refrigerated conditions (Grama et al., 2002).
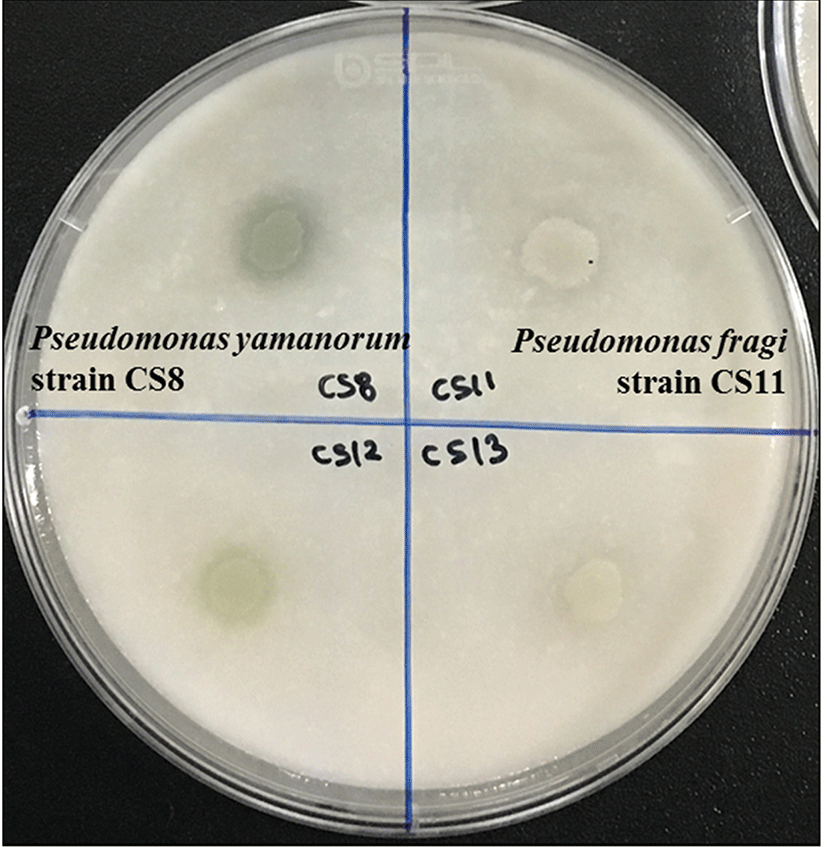
In addition, we demonstrated mixtures of strain-specific characteristics based on proteolytic activity. For example, among 17 strains of Pseudomonas weihenstephanensis tested, two strains had no activity at all, two strains had high activity on chicken juice agar, and the other thirteen strains had moderate proteolytic activity (Table 4). This result was in accordance with other studies that reported different strains exhibited different characteristics (Ercolini et al., 2011; Nieminen et al., 2012).
Conclusion
In this study, we analyzed psychrotrophic bacteria from chicken meat using a culture-dependent approach and next generation sequencing analysis. Though both methods identified similar taxa, the NGS analysis provided much higher resolving power than the cultured method. Therefore, parallel analysis using NGS and culture-dependent approaches provides in-depth information of the biodiversity of the spoilage microbiota in chicken meat.
The predominant genus responsible for chicken meat spoilage was Pseudomonas, but other species belonging to the genera Serratia and Aeromonas were also identified. The combination of psychrotrophic growth of these bacteria in cooled chicken meat with concomitant production of proteolytic enzymes presents a big challenge to the meat industry. Therefore, it is essential to know the dynamics of the microbiota composition as a function of the refrigerated storage time in order to better understand how the specific spoilage bacteria evolves in chicken meat and to develop useful preservation methods.
Consequently, further study is needed to develop better preservation methods against these spoilage bacteria. For example, different packing methods such as modified atmosphere packaging and vacuum packaging should be tested, and bacteriophages targeting specific psychrotrophic bacteria can be applied for this purpose.













