Introduction
Canines are primarily carnivorous animals that naturally consume a meat-based diet (MacDonald et al., 1984). However, in modern times, urban canines have shifted towards high-carbohydrate diets, leading to lifestyles that resemble those of humans. The gastrointestinal (GI) microbiota of these companion animals plays a crucial role in their health and well-being. Imbalances and alterations in the GI microbiota have been closely associated with various GI diseases and disorders in dogs, including diarrhea (Marks et al., 2011) and dysbiosis such as idiopathic inflammatory bowel disease (Suchodolski et al., 2012). As a result, there is a growing interest in understanding and modulating gut microbiota to enhance overall canine health. The composition and function of the GI microbiota in dogs can be influenced by numerous factors, including diet, environmental exposures, and host genetics. Investigating the intricate relationship between the canine microbiota and health outcomes has the potential to uncover novel therapeutic approaches and preventive strategies.
Probiotics, defined as live microorganisms that confer health benefits when consumed in sufficient amounts (FAO and WHO, 2002), represent a promising avenue for optimizing the gut microbiota in dogs. The guidelines established by FAO and WHO (2002), the Food and Drug Administration, and the Ministry of Public Health of Thailand outline the requirements for the use of probiotic microorganisms in food, including accurate identification, determination of probiotic characteristics (such as resistance to gastric acid, bile salt resistance, adherence to the mucosa, epithelial cells, and cell lines, as well as bile salt hydrolase activity), and safety assessment encompassing antimicrobial resistance (AMR), toxin production, and hemolytic activity (Binda et al., 2020; FAO and WHO, 2002; Ministry of Public Health of Thailand, 2022). These guidelines are equally applicable to canine probiotics.
Probiotic supplements have shown health-promoting properties in both healthy and diseased canines. They help regulate the gut microbiota, stimulate immune function, enhance nutrient metabolism, and contribute to the prevention and mitigation of various diseases, including digestive disorders, infectious diseases, cancer, and allergies (Michail et al., 2006). Probiotics isolated from fecal samples of healthy dogs can serve as potent dietary supplements for canines (Sivamaruthi et al., 2021). In recent years, whole-genome sequence analysis has emerged as a valuable tool for the accurate identification and safety evaluation of probiotic products (Soni et al., 2021).
Recognizing the significant role of the GI microbiota in canine health, this study aims to isolate probiotic strains from canine feces and evaluate their potential probiotic properties. The evaluation encompasses a comprehensive range of parameters, including antibacterial activity, tolerance to acid and bile salts, auto- and co-aggregation adhesion, cytotoxicity, hydrophobicity, as well as β-galactosidase and antioxidant activities. Furthermore, whole-genome sequencing will be conducted to explore the presence of genomic determinants related to AMR, prophage elements, clustered regularly interspaced short palindromic repeats (CRISPR), bacteriocin-encoding genes, β-galactosidases, stress responses, cell adhesion, and secondary metabolite-related genes. This analysis will utilize publicly available databases to gain insights into the safety profile of the probiotics in silico. This study represents the first comprehensive investigation into the probiotic species Enterococcus hirae, Limosilactobacillus fermentum, Pediococcus pentosaceus, and Ligilactobacillus animalis, isolated from dogs. The investigation encompasses both phenotypic and genotypic analysis through whole genome sequencing. Thoroughly examining the characteristics of these probiotic strains will contribute to the development of interventions focused on enhancing the GI health of dogs.
Materials and Methods
All strains of lactic acid bacteria (LAB) used in this study were cultivated on de Man, Rogosa, and Sharpe (MRS) agar plates or MRS broth (HiMedia, Mumbai, India) and incubated anaerobically at 37°C. The bacterial strains used for the inhibition tests were Escherichia coli ATCC 25922, Bacillus cereus JCM 2152, Salmonella Typhimurium TISTR 1471, and Staphylococcus aureus ATCC 25923, which were propagated on nutrient agar slants (HiMedia).
Fecal samples were collected from canines after authorization from the owners. Thirty healthy canines of different breeds [pomeranian (Pom), French bulldog (FB), chihuahua (Chi), mongrel canines (MD), Shih Tzu (Shi), and poodle (PD)] were sampled for feces to isolate LAB strains. Research involving the use of animals was conducted in accordance with the guidelines of the Institutional of Animals for Scientific Purposes Development (IAD), Thailand, under the reference number U1-00263-2558. The fecal samples were serially diluted and spread onto MRS agar plates supplemented with 0.1% CaCO3, followed by anaerobic incubation at 37°C for 24–48 h. Colonies exhibiting clear halos were purified and subjected to evaluation for morphological and biochemical characterization, following the method described by Schillinger and Lücke (1987). Amplification of the 16S rDNA was performed using a standard PCR protocol with universal primers 27F (5’-AGAGTTTGATCCTGGCTCAG-3’) and 1492R (5’-TACGGYTACCTTGTTACGACTT-3’; Stackebrandt and Goodfellow, 1991). The PCR products were separated by electrophoresis on a 1% (w/v) agarose gel and visualized after staining with GelRed (Biosharp, Anhui, China). Subsequently, the PCR products were purified, and sequencing was carried out. Similar searches were performed in GenBank using BLAST (http://www.ncbi.nlm.nih.gov/blast).
A bacterial suspension of 1.5×108 CFU/mL (equivalent to McFarland No. 0.5 standard) was prepared, and 1% (v/v) LAB inoculum was added to MRS broth with varying concentrations of oxgall and at different pH levels (3.5 and 4.5). In both experiments, the positive control was prepared using MRS broth at pH 7, without the addition of bile salt. The negative control, on the other hand, consisted of MRS broth without any bacterial inoculation. Growth monitoring was by measurement of absorbance values at OD600 after 24 h of culturing at 37°C (Guo et al., 2010). Growth under unfavorable conditions was indicated as follows: OD equal to negative controls; no growth (–), OD greater than negative control and less than positive control; weak (+), or OD equal to positive control; good growth (++).
E. coli ATCC 25922, B. cereus JCM 2152, Salmonella Typhimurium TISTR 1471, S. aureus ATCC 25923,Latilactobacillus sakei JCM 1157, Lactiplantibacillus plantarum ATCC 8014, Lactococcus lactis JCM 7638, L. lactis subsp. cremoris TUA 1344L, Leuconostoc mesenteroides JCM 6124, P. pentosaceus JCM 5885, P. pentosaceus JCM 5890, and Streptococcus salivarius JCM 57077 were used as indicator strains to determine the antibacterial activity in accordance with the method of Pringsulaka et al. (2012). The level of inhibition of the test strains was interpreted based on the diameter of the zone of inhibition as follows: high (>15 mm, +++), medium (10–15 mm, ++), low (<10 mm, +), and absent (–) (Nair, 2000).
The adhesion of cells to hydrocarbon (hexadecane) was used to determine cell surface hydrophobic properties. The LAB isolates were allowed to grow in MRS broth at 37°C overnight, washed with sterile 0.85% NaCl, harvested, and re-suspended in MRS broth. Approximately 3 mL aliquots of the bacterial suspensions were exposed to 1 mL of hexadecane (Sigma-Aldrich, St. Louis, MO, USA). The hydrophobicity index (HPBI) was calculated using the formula:
where A1 represents the OD600 of the bacterial suspension before mixing with hexadecane, and A2 represents the OD600 of the aqueous phase obtained after thorough mixing with hexadecane and vortexing for 2 min. Isolates exhibiting a HPBI >70% were categorized as strongly hydrophobic, while those with an HPBI ranging from 50% to 70%, and less than 50% were classified as moderately and weakly hydrophobic, respectively. High hydrophobicity indicates a good adhesive capability (Nostro and Cannatelli, 2004).
The MRS broth culture of LAB cells grown for 24 h was collected, washed twice with 0.85% NaCl, resuspended, and diluted to an OD600 of 0.5 (approximately 108 CFU/mL). The bacterial cell suspensions were then vortexed for 10 sec and incubated at 37°C for 5 h. The auto-aggregation percentage was calculated using the formula:
where A20 represents the OD600 at 20 h and A0 represents the OD600 at 0 h (Oh et al., 2018).
LAB strains were cultivated as previously described, while E. coli ATCC 25922, S. aureus ATCC 25923, Salmonella Typhimurium TISTR 1471, and B. cereus JCM 2152 were cultured in nutrient broth for 24 h at 37°C. LAB and pathogenic bacterial suspensions, each with a volume of 15 mL, were mixed and incubated for 2 h without agitation. Control tubes were prepared with 15 mL of suspension for each bacterial strain. After the incubation period, the absorbances (OD600) of both the mixtures and the controls were measured. The co-aggregation percentage was determined using the following formula:
where Aprobiotic and Apathogens represent the OD600 of the LAB and pathogen cell suspensions, respectively, and Amix represents the OD600 of the bacterial suspension mixture after 2 h incubation (Oh et al., 2018).
Colorectal adenocarcinoma (Caco-2; ATCC HTB-37) cells were seed at a density of 1×105 cells/mL in 24-well plates and cultured at 37°C in a humidified atmosphere containing 5% CO2 prior to the adhesion assays. LAB pellets obtained by centrifugation at 10,000×g were seeded in a 24-well plate at 105 CFU/mL per well and further incubated at 37°C for 4 h. Subsequently, the bacteria were aspirated, and the wells were rinsed with PBS. Next, the wells were treated with 0.5% Triton X-100 to facilitate the separation of bacteria. The bacterial count was determined on MRS agar, and the adhesion rate (%) was calculated according to the following equation:
where N represents the CFU of probiotic bacteria after adhesion to the Caco-2 cell line for 4 h, and N0 represents the CFU the probiotic bacteria that were initially inoculated (Jang et al., 2019).
The cytotoxic activities of the four probiotic cell extracts were determined following the method described by Awaisheh et al. (2016). The probiotic cells were cultured in MRS medium at 37°C for 24 h, and the supernatant was collected by centrifugation at 10,000×g for 5 min. Subsequently, the resulting supernatant was filtered through a 0.45-μm filter membrane to obtain the filtered portion, which was used for further testing.
Vero cells were seeded at a density of 2×103 cells/well in a 96-well microplate. The cells were then treated with filter-sterilized supernatant of the isolated strain and further incubated in a humidified environment containing 5% CO2 for another 24 h at 37°C. After treatment, the medium was replaced with 20 μL solution of 3-(4,5-dimethylthiasol-2-yl)-2,5-diphenyltetrazolium bromide (MTT; Merck, Rahway, NJ, USA) along with 180 μL of completed DMEM. The cells were then further incubated for 4 h at 37°C. Subsequently, the MTT mixture was carefully discarded, and 0.1 mL of dimethyl sulfoxide (Merck) was added. The absorbance was measured at 570 nm. Untreated cells were used as controls. Cytotoxicity was calculated according to the following equation:
The safety of the selected isolates was evaluated based on their hemolytic activity on Columbia agar supplemented with 5% (v/v) sheep blood, as described by Lombardi et al. (2004). Each isolate was streaked on the agar in three replicates. After incubation at 37°C for 48 h, the plates were observed for the presence of hemolytic reactions. Alpha-hemolysis is indicated by the formation of a green zone around the colonies, which indicates partial hemolysis. Beta-hemolysis is indicated by the formation of a clear (transparent) zone around the colonies, which indicates complete hemolysis. Gamma-hemolysis is indicated by no change in the medium, indicating no hemolysis occurred.
The antibiotic susceptibility of the LAB strains was determined using a MIC Test Strip (Liofilchem® MTSTM, Roseto degli Abruzzi, Italy). Overnight cultures of LAB were adjusted to 0.5 McFarland standard and diluted to 5×105 CFU/mL. Subsequently, 0.1 mL of the LAB suspension was plated onto Mueller–Hinton agar plates (HiMedia). The MIC Test Strips containing ampicillin, chloramphenicol, erythromycin, gentamicin, and tetracycline (Liofilchem® MTSTM) were positioned at the center of the plate and incubated for 24 h at 37°C. These antibiotics were chosen based on their inclusion in the European Food Safety Authority (EFSA) list. The MIC of each antibiotic was determined by evaluating the ellipsoid zones of inhibition of bacterial growth and determining the point of intersection between these zones and the concentration mark on the test strip. Susceptibility or resistance was assessed in accordance with the microbial cutoff values recommended by EFSA (2012).
The biogenic amines production was examined using decarboxylase base medium supplemented with 2% histidine, lysine, ornithine, and tyrosine (Sigma-Aldrich). The base medium was incorporated into MRS broth at a concentration of 0.25% (w/v). After incubation for 4 days at 37°C, the presence of a purple halo was considered as a positive reaction (Joosten and Northolt, 1989).
DPPH free radical activity of the isolates was determined as described by Das and Goyal (2015). Overnight LAB cell suspension (2 mL, 109 CFU/mL) was mixed with 2 mL of 0.4 mM DPPH solution (Sigma-Aldrich) in 99.8% methanol, and the mixture was vortexed for 5 min. Subsequently, the mixture was incubated in the dark for 30 min. The samples were subjected to centrifugation at 8,000×g for 10 min and the absorbance was then measured at 517 nm. Ascorbic acid (1 mg/mL) was employed as the positive control. The DPPH scavenging activity was calculated using the following formula:
where Asample represents the OD517 of the mixture of bacterial cells and DPPH solution, Ablank represents the OD517 of the mixture of methanol and bacterial cells, and Acontrol represents the absorbance of the DPPH solution.
β-Galactosidase activity was determined using a method described by Chen et al. (2002), with minor modifications. Overnight cultures were collected by centrifugation, followed by washing with PBS buffer (pH 7.0). The bacterial cells were then added to Z-buffer (60 mM Na2HPO4, 40 mM NaH2PO4, and 2.7 μL/mL β-mercaptoethanol), and absorbance (A600) was measured. Next, 0.1 mL of the cell suspension was mixed with 0.9 mL Z-buffer and 0.02 mL toluene, vortexed, and incubated for 1 h. Then, 0.2 mL of 200 mM ONPG solution (Sigma-Aldrich) was added and further incubated for 30 min. Next, 0.5 mL of 1 M Na2CO3 was added to stop the reaction. The absorbances at A420 and A560 were measured. The activity of β-galactosidase (Miller units) was calculated using the following formula:
where A600 represents the absorbance of cells before the assay, A550 represents the absorbance of cell debris after the assay, A420 represents the absorbance of o-nitrophenol (ONP) released, T is the reaction time (min). V is the volume of culture used (mL).
Genomic DNA of the LAB strains was extracted using an AccuPrep® Genomic DNA Extraction Kit (Bioneer, Daejeon, Korea), following the manufacturer’s instructions. The DNA concentration and quality were assessed using a NanoDropTM 2000 spectrophotometer (Thermo Fisher Scientific, Waltham, MA, USA). The purified genomic DNA was submitted to the Beijing Genomics Institute (BGI) in China for short-read sequencing. Next, 1 μg of qualified genomic DNA was randomly fragmented using Covaris. Fragments of 800 bp were selected using the Agencourt AMPure XP-Medium kit. End repair and 30-adenylation were performed on the fragments, followed by the ligation of adaptors to the ends of these 30-adenylated fragments for amplification. Subsequently, the PCR products were purified using an Agencourt AMPure XP-medium kit. Splint oligo sequences were used to heat denature and circularise the double-stranded PCR products. Single-stranded circular DNA was used as the final DNA library.
De novo assemblies of the LAB genome sequences were constructed using SPAdes v3.12 (Bankevich et al., 2012). The quality and completeness of the genome assemblies were evaluated using Quast v5.0.2 (Gurevich et al., 2013), and genome annotation was performed using Prokka v1.12 (Seemann, 2014).
AMR genes, plasmids, prophages, and virulence-related genes were identified to assess the safety of selected probiotic strains. The AMR genes were identified using ResFinder v2.1 with a 90% identity threshold and 60% minimum coverage. Furthermore, the genome stability was evaluated using several pipelines, including PlasmidFinder (Carattoli and Hasman, 2020), PathogenFinder (Cosentino et al., 2013), and the virulence factors database (VFDB, http://www.mgc.ac.cn/VFs/main.htm) with a cut-off of >75% identity and >60% coverage. The completeness of the predicted phage-related regions was determined using PHASTER, which classified them as intact (>90%), questionable (90%–60%), or incomplete (<60%) regions based on the number of known genes and proteins contained in the bacterial prophage region (Arndt et al., 2016).
CRISPR and bacteriocin-encoding genes were identified using the CRISPRFinder (Grissa et al., 2007) and BAGEL4 web servers, respectively (van Heel et al., 2018). Genomic data were visualized using CGView Server v1.0.
Results and Discussion
Twenty LAB isolates that exhibited gram-positive characteristics and tested negative for both catalase and oxidase were selected. These isolates displayed distinct morphologies, with 6 isolates showing a spherical shape, 8 isolates showing a rod shape, and 6 isolates showing an oval shape. Subsequently, the colonies underwent additional in vitro screening to assess their probiotic characteristics.
A prerequisite for probiotics is their ability to resist harsh conditions in the stomach and small intestine (Ouwehand et al., 2003). Therefore, all 20 LAB isolates were assessed for their survivability in acidic conditions (pH 3.5, 4.5) and in the presence of 0.3%, 0.5%, and 1.0% bile salts (Table 1). Most of the isolates showed tolerance to pH 3.5. However, at pH 4.5, only 9 isolates, namely FB2, Pom1, Pom4, Pom5, Pom9, Chi5, Chi8, MD3, and MD12, exhibited growth comparable to the control without pH adjustment. Nearly all of them exhibited growth in the presence of 1.0% bile salts. These findings can be attributed to the exposure of these isolates to low pH and bile salts in the GI tract, as they are commonly found in animal feces. However, the tolerance levels of these isolates to acidic environments varied. This is consistent with the results reported by Kumar et al. (2017), where 9 isolates from canine feces displayed robust growth at pH 4, moderate growth at pH 6, and weak growth at pH 2. Furthermore, all the isolates demonstrated resistance to bile at a concentration of 0.3% oxgall. However, there was a decrease in CFU count for all isolates at the 1% bile salt concentration.
To evaluate the hydrophobic properties of the LAB strains, we measured their microbial adhesion to solvents. As shown in Table 1, isolates Pom1 (88.91%) and Pom2 (88.45%) demonstrated the highest degree of hydrophobicity towards hexadecane. It is worth mentioning that a previous study on the isolation of probiotics from canine feces reported a percent hydrophobicity exceeding 80% for Lactobacillus johnsonii cPRO23 (Kumar et al., 2017). However, it is important to consider that the solvents used in that study were toluene and xylene, which differ from the solvent (hexadecane) employed in our investigation. Previous studies have reported that LAB with a higher hydrophobicity of their cell surface may exhibit greater adhesion to Caco-2 cell lines. For instance, Krausova et al. (2019) investigated the cell surface hydrophobicity of 19 strains of L. fermentum and Lactobacillus casei, finding that the hydrophobicity values ranged from 0.3% to 68.8%. It is important to note that the variations in cell surface hydrophobicity may stem from different methods used to assess adhesion, such as the Bacterial Adherence to Hydrocarbons (BATH) method, which involves testing adhesion to hydrocarbon compounds. Other factors, including the duration of incubation, composition of the growth medium, and type of hydrocarbon compounds used, can also contribute to these variations.
The inhibitory activity of all isolates was assessed against selected Gram-positive and Gram-negative bacteria. In the agar-well diffusion test, the LAB isolates exhibited varying degrees of inhibitory activity against the indicator strains, with inhibition zones ranging from 11 to 15 mm, as shown in Table 1. Out of the tested strains, 60% (12/20) exhibited inhibitory activity against B. cereus. However, only 15% (3/20) of the isolates demonstrated activity against Salmonella Typhimurium. Additionally, among the tested indicators, only three isolates (Pom4, Pom5, and Chi8) showed activity against all four indicators. None of the isolates produced bacteriocin when the different indicator microorganisms were used; however, the three putative bacteriocins open reading frames (ORFs), enterolysin A, hiracin, and class II lanthipeptide were mined from the genome of E. hirae Pom4 (data not shown).
The auto-aggregation ability is especially crucial as it enables the probiotics to form cellular aggregates, which indicates their capacity to colonize the intestine. Furthermore, co-aggregation of probiotics is critical for prevention of surface colonization of pathogenic strains (Collado et al., 2008). The auto-aggregation and co-aggregation abilities of the probiotics against foodborne pathogens were studied by measuring the percentage of aggregation after 20 h of incubation at 37°C. Out of the tested LAB strains, FB1, FB2, and Chi3 showed the highest auto-aggregation abilities (80.76 0.08%, 86.98 0.12%, and 81.75 0.14%, respectively). The co-aggregation of probiotic strains and E. coli, S. aureus, Salmonella Typhimurium, and B. cereus is shown in Table 1. Co-aggregation of probiotics is critical for prevention of surface colonization of pathogenic strains. Among the LAB strains evaluated, FB2 showed the highest co-aggregation ability with E. coli (82.03 0.10%), Salmonella Typhimurium, (82.28 0.15%), S. aureus (82.63 0.12%), and B. cereus (85.81 0.41%). Among the pathogens, B. cereus and S. aureus demonstrated the highest auto-aggregation abilities. This implies that the probiotic bacteria can proficiently bind with each other, or other bacteria present in the gut. Such a capability can potentially boost the growth of beneficial gut bacteria, diminish the growth of harmful bacteria, and aid immune function. In a previous report, it was observed that LAB isolated from canines exhibited co-aggregation percentages with Salmonella Typhimurium ranging from 35% to 45%. This percentage was higher than that of the reference strain L. acidophilus NCDC 15, which was isolated from a dairy source.
The 20 LAB strains demonstrated potential probiotic characteristics, including robust pH tolerance, bile tolerance, and the ability to adhere to the intestinal mucosa, which indirectly indicates hydrophobic properties. These LAB were subjected to 16S rDNA sequence analysis. The isolates, Chi4, and Chi5 were identified as Enterococcus faecium; FB2 as L. animalis; Pom4, Chi3, and MD1 as E. hirae; Pom5, MD3, MD12, and Shi1 as L. fermentum; Chi6 as Enterococcus avium; Chi7 and Chi8 as P. pentosaceus; MD2 as Streptococcus lutetiensis; MD13 and PD3 as E. faecalis; and Pom1, Pom2, and Pom9 as Lactobacillus sp. (Supplementary Table S1).
The present study demonstrated that four representative LAB strains (E. hirae Pom4, L. animalis FB2, L. fermentum Pom5, and P. pentosaceus Chi8) possess significant probiotic properties. These strains are nonpathogenic, exhibit resilience to acid and bile salts, demonstrate a broad spectrum of antibacterial activity against pathogenic strains, adhere to cell surfaces, and show robust growth in MRS broth within a 48-h period, making them well-suited for further experimental cultivation. Consequently, the present study evaluated the adhesion ability of these strains, and it was found that all of them exhibited varying degrees of adherence to Caco-2 cells. Among the strains evaluated, P. pentosaceus Chi8 displayed the highest adhesion capacity (95.08±2.92%), followed by L. fermentum Pom5 (88.40±1.85%), L. animalis FB2 (83.95±1.70%), and E. hirae Pom4 (76.92±0.56%). These results suggest that adhesion is a strain-specific property. The capacity to adhere is crucial for transient colonization, antagonism against pathogens, modulation of the immune system, and promoting healing of damaged gastric mucosa (Alander et al., 1999). The LAB isolates from canines were found to exhibit binding to human mucosa, consistent with their binding ability to canine mucosa.
The cytotoxicity of the four strains was evaluated against the Vero cell line using an MTT assay. After incubation for 4 h with cell-free supernatant (CFS) of the respective strains, the cell viability was 74.58%–93.26% (data not shown). The CFS of the four probiotic strains was harmless to the noncancerous-Vero kidney cell line, with L. fermentum Pom5 exhibiting the highest cell viability.
The hemolytic activity and biogenic amine production of LAB isolates were pre-evaluated to confirm that these probiotics are safe to use. None of the selected isolates showed any hemolytic activity (gamma-hemolysis) in the present study. Biogenic amines, when present in high amounts, can be toxic to animals. However, it was determined that the LAB isolates in this study lacked the ability to convert tyrosine, lysine, ornithine, and histidine into tyramine, cadaverine, putrescine, and histamine, respectively. This finding indicates that these isolates are considered safe for canine health.
One of the properties required for specific strains to be considered potential probiotics is the absence of acquired and transferable antibiotic resistance (Courvalin, 2006). Therefore, microbes need to be effectively screened for antibiotic resistance genes before their use as probiotics. Strains were considered resistant when they showed values greater than the MIC breakpoints established by EFSA (2012). All four strains were sensitive to ampicillin, chloramphenicol, gentamicin, and erythromycin. However, L. animalis FB2 and E. hirae Pom4 were resistant to tetracycline (data not shown). Generally, LAB is sensitive to broad-spectrum antibiotics, such as tetracycline, chloramphenicol, and beta-lactams. The most frequently observed resistance genes are for tetracycline and erythromycin resistance, followed by those for chloramphenicol resistance (Çataloluk and Gogebakan, 2004).
The DPPH radical scavenging activities of the four LAB strains were as follows: L. fermentum Pom5 (36.39%), E. hirae Pom4 (28.87%), L. animalis FB2 (23.34%), and P. pentosaceus Chi8 (16.42%). These results demonstrate that the scavenging activity of LAB isolates is strain dependent.
Lactose intolerance refers to the discomfort that arises after consuming milk and dairy products due to insufficient amounts of β-galactosidase (lactase) for lactose digestion in the intestine. The production of β-galactosidase by probiotic strains has been proposed as a potential remedy for alleviating the symptoms of lactose intolerance (Ljungh and Wadström, 2006). The four selected strains showed β-galactosidase activity as follows: P. pentosaceus Chi8 (373.48±0.16 Miller unit), L. animalis FB2 (364.23±0.02 Miller unit), L. fermentum Pom5 (327.75±0.18 Miller unit), and E. hirae Pom4 (301.90±0.01 Miller unit).
A positive correlation was observed between lactose sugar utilization and the genome. A lactose utilization gene cassette was found in the genomes of all four LAB strains, including lacLM, and lacS, which encode β-galactosidase and lactose permease, respectively. Additionally, lacR, which encodes a lactose transport regulator, was found in all strains, except E. hirae Pom4. Even though these strains originated from canine intestines, they still produced β-galactosidase. Therefore, in addition to their established probiotic properties, these strains would improve the digestion of milk and dairy products consumed by canines.
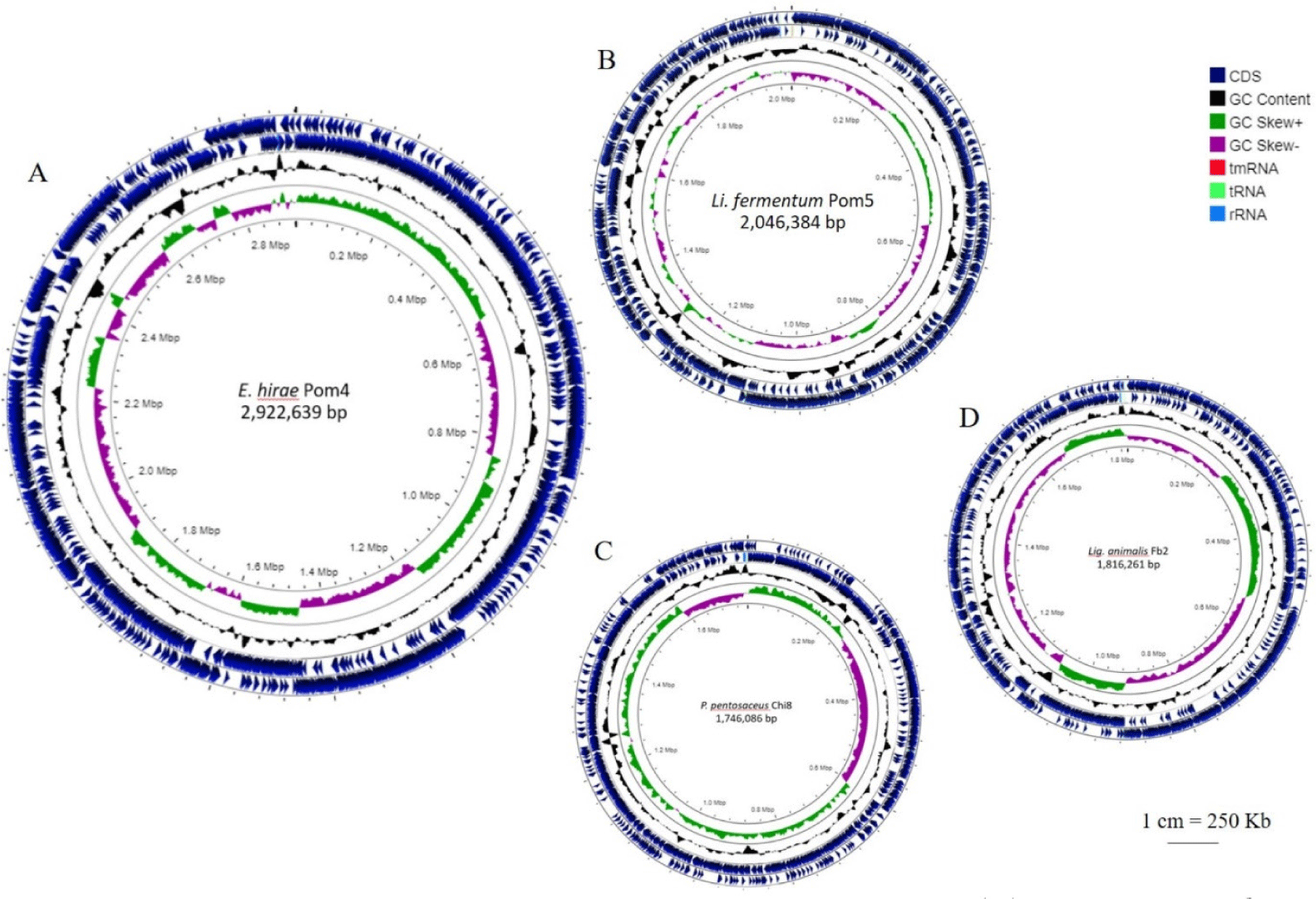
The general genomic features of the four LAB strains are presented in Table 2 and circular representations of the genomes are shown in Fig. 1. Their genome sizes were approximately 1.74–2.92 Mb; P. pentosaceus Chi8 had the smallest genome, whereas E. hirae Pom4 had the largest (Fig. 1). The G+C content varied between 36.6%–52.0%.
The entire genomes of the four selected LAB strains were screened following the recommendations of the EFSA guidelines to search for AMR, virulence factors (VFs), toxin-related genes, and mobile genetic elements (MGEs) to ensure the safety of probiotic bacteria when used as a dietary supplement. The search was performed using two AMR databases, CARD, and ResFinder. The primary concern regarding antibiotic resistance genes (AMR) in beneficial nonpathogenic bacteria is the potential transfer to other potentially pathogenic bacteria, which can result in complications and reduce the effectiveness of antibiotic treatment. To assess this risk, we specifically investigated two types of MGEs, plasmids, and bacteriophages, as they are known to play a role in intercellular genetic exchange through processes such as transformation, conjugation, and transduction. However, it is still necessary to clarify how these AMR genes are acquired and where they are located. If AMR genes are located in MGEs such as plasmids, they can be transferred to other pathogenic species.
This study focused on transferable antibiotic-resistance or acquired-AMR genes because probiotics may serve as a reservoir for the potential spread of resistance genes among bacteria. No AMR genes were identified in the genomes of L. fermentum Pom5, and P. pentodaceus Chi8 using ResFinder; this suggests that they are safe probiotics. In contrast, AMR genes were detected in E. hirae Pom4 and L. animalis FB2 (Supplementary Table S2). Tetracycline is one of the most frequently used antibiotics for human and animal infections, owing to its availability and low cost. The detection of tetM and tetL in the genome of E. hirae Pom4 is consistent with previous reports that tetL, which encodes an efflux pump protein, does not appear alone in LAB but is always detected together with other tetracycline resistance genes. There are two fundamental mechanisms of resistance to tetracyclines in enterococci, flow pumps and ribosome protection, which prevent antibiotic binding (Miller et al., 2014). Additionally, aminoglycoside 6’-N-acetyltransferase (aac(6’)-Ian), which is responsible for aminoglycoside resistance, was found in E. hirae Pom4 with 100% identity (Supplementary Table S2). Different plasmid-associated replication genes ensuring genome stability were found only in E. hirae Pom4 and P. pentosaceus Chi8 (Supplementary Table S3). This indicates that the plasmids may have been inserted into the genomes of E. hirae Pom4 and P. pentosaceus Chi8. In E. hirae Pom4, tetracycline resistance genes were found in the same region as the plasmid-related replication genes. Horizontal gene transfer is relevant for disseminating antibiotic resistance in non-human hosts, with plasmids playing a central role in this process. However, P. pentosaceus Chi8 did not exhibit any antibiotic resistance genes in the region of plasmid-associated replication genes. According to the safety guidelines for probiotics, E. hirae Pom4 is not a suitable probiotic candidate, despite it possessing several desirable properties. Numerous commercially available probiotic products for companion animal consumption include enterococci, the natural flora of canine and feline GI tracts. Some strains can exert their beneficial effects on the host as probiotics, while others can spread antibiotic resistance to other bacterial cells. Therefore, although no probiotic-enterococcal infections have been reported in animals, they pose certain theoretical safety risks. As such, the antibiotic resistance of these bacteria must be carefully evaluated before they are used as commercial probiotics. In the case of L. animalis FB2, only a tetracycline resistance gene, tetM, was detected in the chromosomal regions, representing a low risk of transferring the antibiotic resistance gene.
MGEs, such as prophages, transposases, gene islands, and insertion elements, play a major role in bacterial horizontal gene transfer. Prophages, in particular, may play a crucial role in lateral gene transfer among strains, contributing to genetic diversity and strain specificity. These factors can be used to assess genomic diversity during bacterial evolution (Liu et al., 2022). The presence of prophage sequences in each of the four evaluated probiotics was analyzed using the PHASTER web server. Intact prophages carry all the necessary genes for excision and reinfection, whereas incomplete prophages lack some of these genes, indicating their non-functionality. E. hirae Pom4 and P. pentosaceus Chi8 carried a single intact prophage region. However, several incomplete prophage regions were identified in L. fermentum Pom5, P. pentosaceus Chi8, and L. animalis FB2 (Supplementary Table S4).
The PathogenFinder web server predicted that L. fermentum Pom5 and P. pentosaceus Chi8 were non-human pathogens (probability, 0.177). On the other hand, E. hirae Pom4 and L. animalis FB2 were predicted to be human pathogens with probabilities of 0.798 and 0.64, respectively. A BLAST search against VFDB was performed to determine the presence of VFs within the genomes of all four selected LAB strains. No virulence genes were identified in L. fermentum Pom5, P. pentosaceus Chi8, and L. animalis FB2. However, two notable virulence determinants were identified in the genome of E. hirae Pom4, bopD (maltose operon transcriptional repressor MalR, LacI family, and biofilm formation proteins) and clpP (proteolytic subunit of ATP-dependent Clp protease). These genes were identified as VFs in VFDB and are also implicated in the adaptation, survival, and attachment of pathogenic bacteria to adverse environments. The absence of virulence determinants in E. hirae Pom4 and P. pentosaceus Chi8 represents a precondition for their consideration as potential probiotics.
CRISPRs are genetic elements that are formed by the repetition of DNA sequences within a specific genomic area. CRISPRs and their associated cas genes play important roles in defending organisms against invasive MGEs (Marraffini and Sontheimer, 2008). The CRISPRFinder and BAGEL4 web servers were used to identify a known type of CRISPR region and Cas cluster in L. fermentum Pom5 and L. animalis FB2. However, an unknown type of CRISPR region was found in E. hirae Pom4 (two regions) and P. pentosaceus Chi8 (one region). The presence of the CRISPR/Cas system within these four probiotic strains demonstrates the stability of their bacterial genomes and immunity against the spread of acquired AMR genes by obstruction of multiple pathways involved in horizontal gene transfer.
Several stress response genes were identified in the genomes of all four strains using subclass system analysis of the Pathosystems Resource Integration Center (PATRIC). Genes encoding proteins involved in stress response encoded in the genome of E. hirae Pom4, L. fermentum Pom5, L. animalis FB2, and P. pentosaceus Chi8 are shown in Supplementary Table S5. Moreover, several genes associated with acid and bile resistance were identified. Membrane-bound ATP synthases (F0F1-ATPases) serve as primary regulators of cellular pH inside the cell. Under stress conditions, ATP synthases function as ATPases, thereby generating a transmembrane ion gradient at the expense of ATP hydrolysis (Liu et al., 2015). Bile salt tolerance is another essential property required for the survival of probiotic candidates in the small intestine. The bile salt tolerance capacity of candidate probiotic strains was confirmed by detecting ORFs that encode choloylglycine and bile acid hydrolases, glycine betaine ABC transport system, and ornithine decarboxylase in the genome. Additionally, a cluster of genes related to temperature stress (hrcA-grpE-dnaK-dnaJ and GroEL-GroES), which protect proteins against improper folding and aggregation, were identified. Furthermore, the adaptation of probiotic bacteria to high osmolarity environments is explained by the presence of genes encoding the ABC transporter permease and glycerol uptake facilitator proteins, which are involved in the accumulation of compatible solutes such as proline and glycine betaine (Soni et al., 2021). We further identified the following DNA repair and protection genes: DNA repair protein (MutHLS, Rec and UvrABCD system), ImpB/MucB/SamB family protein, exodeoxyribonuclease, and SbcCD exonuclease. In addition, these findings suggest that the four probiotic bacterial strains effectively respond to DNA damage from stressful conditions.
The probiotics’ capability to adhere to the intestinal mucosa and epithelial cells was confirmed by the presence of genes related to adhesion, colonization, fibrinogen/fibronectin binding, enolase, and glyceraldehyde-3-phosphate dehydrogenase (Supplementary Table S6). These adhesion genes may help the strains to exert probiotic effects. For example, mucin and fibrinogen are adhesion proteins that facilitate probiotics in binding to the digestive tract, enhancing colonization and reducing pathogenic adhesion (Granato et al., 2004). Additionally, genes related to aggregation were also found in the candidate probiotic strains, such as LysM pepticaninelycan-binding domain-containing protein, translation elongation factor Tu GroEL chaperone, and peptidyl-propyl cis-trans isomerase. Moreover, proteins associated with exopolysaccharide biosynthesis, including tyrosine-protein kinase transmembrane modulator, UTP--glucose-1-phosphate uridylyltransferase, and glycosyltransferase, were detected in the candidate probiotic strains.
The genomes of the candidate probiotic strains were found to contain functionally active biosynthetic genes that encode proteins related to the synthesis of vitamins and essential amino acids (Supplementary Table S7). Genome analysis revealed that L. fermentum Pom5 carries several genes that are involved in the synthesis of vitamins B1 (thiamine), B2 (riboflavin), B6 (pyridoxine), B7 (biotin), and B9 (folate), and essential amino acids, such as arginine, histidine, lysine, phenylalanine, and threonine. This finding demonstrates that in addition to its potent probiotic activity, L. fermentum Pom5 also produces essential nutrients that may be beneficial for canine health.
Conclusion
This study examined the traits of four candidate probiotic strains isolated from canine fecal samples in Thailand. Owing to the variation in canine GI microbiota in different areas, potential probiotic candidates were screened for probiotic attributes and evaluated for safety properties through a combination of genome analyses and phenotypic tests. According to EFSA guidelines, genome analyses indicated that L. fermentum Pom5 and P. pentosaceus Chi8 possessed characteristics of safe probiotic strains, including the absence of transferable antibiotic-resistant and VF genes, and genome stability. Additionally, genes coding for proteins responsible for survival under gastric conditions, including stress response, acid and bile tolerance, and adhesion proteins, enhanced the probiotic properties of the candidate strains. These probiotics may affect canine metabolism by synthesizing various essential amino acids such as arginine, histidine, lysine, phenylalanine, threonine, and B group of vitamins. The selected strains were among the host-specific LAB isolated from canines, which could serve as potential probiotics for canines, particularly in Thailand, where all products are imported.













