Introduction
Escherichia coli is a gram-negative, rod-shaped bacteria present in the guts of various animals, including mammals (Jang et al., 2017). Most strains of E. coli are non-pathogenic but can cause opportunistic infections (De Souza et al., 2019), and some highly pathogenic strains cause various intestinal diseases and sepsis in humans and animals (Fabian et al., 2020).
E. coli subtypes include enterotoxigenic E. coli (ETEC), enteropathogenic E. coli (EPEC), enterohemorrhagic E. coli (EHEC), enteroaggregative E. coli (EAEC), and enteroinvasive E. coli (EIEC) (Al-Gallas et al., 2007). ETEC frequently infects newly weaned pigs, causing acute diarrhea and even death. It can also cause diarrhea, abdominal pain, and dehydration in humans (Pattabiraman et al., 2018). A recent study showed that infections with ETEC delay the growth of pigs and cause mortality owing to excessive diarrhea, resulting in huge economic losses (Yan et al., 2009). To prevent and treat ETEC in pigs, many antibiotics such as oxytetracycline and spectinomycin are commonly administered. However, excessive use of antibiotics can cause negative side effects, such as drug resistance and drug residues in pig meat (Luppi, 2017). Thus, the use of antibiotics tends to be limited worldwide, and safe biological alternatives are needed (Laxminarayan et al., 2013).
It has been common to use antibiotics in animal feed to prevent animals from disease and improve production performance in animal husbandry. On the one hand, the emergence of antibiotic-resistant bacteria due to the unreasonable use of antibiotics has sparked efforts to develop alternatives to antibiotics (Cheng et al., 2014). Bacteriophages (also known as phage) are bacterial viruses that can specifically infect bacteria and replicate in host bacteria (Chang et al., 2019). The phages are attracting attention as an alternative to antibiotics because they have a low risk of remaining in the intestine and can evade the resistance developed to antibiotics through different mechanisms (Kebriaei et al., 2020; Lee et al., 2017). Phage can also be an environmentally friendly approach to control infection and contamination of pathogens in foods of animal origin, including meat and poultry products (Bao et al., 2015). ListShieldTM (Intralytix, Baltimore, MD, USA) is a phage-containing food additive that is a non-chemical antibacterial agent to control the food-borne bacterial pathogen Listeria monocytogenes (Gutiérrez et al., 2017). The ListShieldTM was first approved by the FDA in 2006 as a spray disinfectant that inhibits Listeria growth by applying phage to ready-to-eat deli meats (Hesse and Adhya, 2019). The first GRAS (generally recognized as safe) designation for phage products suggested the possibility of phage food application (Bai et al., 2016). Using phage as a food additive can be an effective method for establishing a safety management system for foods of animal origin but requires in-depth research and approach in terms of safety (Endersen et al., 2014).
In this study, phage ØCJ19 infecting ETEC was isolated from porcine feces, and biological characteristics were evaluated to demonstrate the safety and possibility of phage application to foods of animal origin. Additionally, the whole genome sequence was annotated, and phage ØCJ19 was evaluated as a biological agent to control ETEC.
Materials and Methods
Fecal samples were obtained from a pig farm in Hongseong-gun, Chungcheongnam-do, Korea, suspended in 50 mL SM buffer (50 mM Tris-HCl [pH 7.5], 0.1 M NaCl, and 8 mM MgSO4·7H2O), and centrifuged at 4,000×g for 10 min. The supernatant was filtered using a 0.45 μm membrane filter (Dismic®-25CS, Advantec, Japan) to prepare a sample solution. Next, 18 mL supernatant and 2 mL ETEC FC02 bacterial cell culture (OD540=0.4) were mixed and incubated at 37°C for 18 h. After centrifugation at 4,000×g for 10 min, the supernatant was filtered using a 0.45 μm membrane filter. Then, 100 μL filtered supernatant was mixed with the host bacteria E. coli FC02 bacterial cell culture and 5 mL Luria-Bertani (LB) broth containing 0.7% soft agar. The mixture was poured into 1.5% LB agar and incubated overnight at 37°C. Individual single plaques were harvested from the agar plate using sterile pipette tips, resuspended in 500 μL SM buffer, and the double-layer agar technique was repeated (Šimoliūnas et al., 2014).
A single colony of E. coli FC02 was resuspended in 100 mL LB broth and incubated with vigorous shaking at 37°C. When the culture reached OD540=0.4, it was centrifuged at 4,000×g for 10 min. The bacterial pellets were collected and resuspended in 3 mL SM buffer. The phage lysates were added to the suspension at multiplicity of infection (MOI) of 0.1 and mixed rapidly. After incubation for 20 min at 37°C with intermittent shaking, the suspension was added to 500 mL LB broth and incubated overnight. Chloroform was added to a concentration of 2%, and the culture was further incubated at 37°C with shaking for 30 min. Next, the culture was centrifuged at 11,000×g for 10 min at 4°C, and the supernatant was filtered using a 0.45 μm membrane filter. Solid polyethylene glycol (PEG 8000) was added to the suspension to a final concentration of 10% (w/v), and the mixture was cooled on ice for 2 h. The lysate was centrifuged at 11,000×g for 100 min, and the supernatant was discarded. The pellet was resuspended in 8 mL SM buffer, and the same volume of chloroform was added. The mixture was vortexed and then centrifuged at 3,000×g for 15 min at 4°C. The supernatant containing the phage particles was recovered and filtered using a membrane filter (Alves et al., 2014).
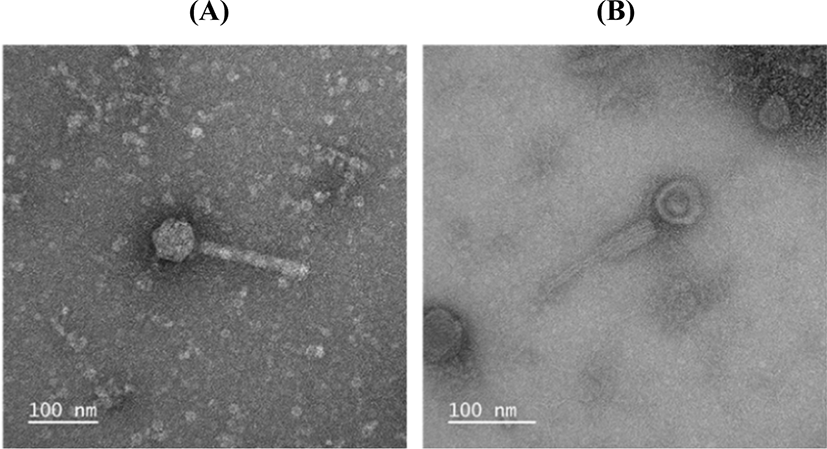
The morphology of phage ØCJ19 was examined using TEM (JEM-2100, JEOL, Tokyo, Japan) at 200 kV. A high-titer phage solution containing 1×108 plaque-forming unit (pfu)/mL was negatively stained with 2% (w/v) uranyl acetate. The phage particles were placed on a carbon coating grid and dipped into distilled water containing a drop of 2% uranyl acetate. The 200-mesh grids (Gatan, Pleasanton, CA, USA) were coated with a collodion film prepared from 2% collodion in amyl acetate and used to absorb carbon film fragments with phage particles. After air drying for 10 min, the grids were subjected to TEM. Images of the negatively stained phage particles were obtained using a one-view camera (Gatan) at ×100,000 and ×150,000 magnification (Kim et al., 2018).
Growth curve of E. coli FC02 for determining phage infection time. Bacterial cultures were grown in LB broth in a shaking incubator (200 rpm) at 37°C for 9 h. Measurements of absorbance (optical density, OD=540 nm) were performed every 1 h.
For one-step growth curve analysis, 5 mL E. coli FC02 culture in the exponential phase (OD540=0.4) was centrifuged at 4,000×g for 10 min. The pellet was resuspended in 1 mL LB broth. The phage solution was added to the suspension at a MOI of 0.1. The mixture was incubated at 37°C for 30 min and then centrifuged at 11,000×g for 10 min. The pellet was resuspended in 10 mL fresh LB broth and incubated at 37°C. The phage concentration in the solution was measured at 5 min intervals for 30 min using the double-layer agar technique (Cao et al., 2015). Burst size was calculated as phage out/phage in.
To determine the stability of the phage at various temperatures, 1 mL phage suspension containing 1×108 pfu/mL was incubated at 4°C, 15°C, 25°C, 37°C, 45°C, 55°C, 65°C, 75°C, or 80°C for 2 h. The phage concentration was then measured using a double-layer agar technique. To analyze the stability of the phage at various pH levels, the pH of the SM buffer was adjusted to various levels between pH 2 and 13. A 100 μL phage suspension containing 1×109 pfu/mL was inoculated in 900 μL pH-adjusted SM buffer to obtain a final concentration of 1×108 pfu/mL. After incubation at 37°C for 2 h, the phage concentrations were measured using the double-layer agar technique (Kim et al., 2019). This experimental procedure was performed three times. The results are reported as means±SD from three observations.
Phage genomic DNA was purified using the Phage DNA Isolation Kit (Norgen Biotek, ON, Canada) and subjected to next-generation sequencing (NGS) using a HiSeq 4000 instrument (Illumina, San Diego, CA, USA). A DNA fragment library was constructed using the TruSeq DNA PCR Free Library Preparation Kit (Illumina, San Diego, CA, USA). A de novo assembly generated in the SOAPdenovo and SNAPGENE software (GSL Biotech; https://www.snapgene.com) was used for open reading frames (ORFs) prediction for the whole genome (Mohammed and Cormican, 2015). The genome sequence was compared with other genomes in GenBank via the BLAST program (http://blast.ncbi.nlm.nih.gov/). The whole genome sequences of phages ØCJ19 (GenBank accession No. MT176427) and ØCJ20 (GenBank accession No. MT533174) were deposited in the NCBI database.
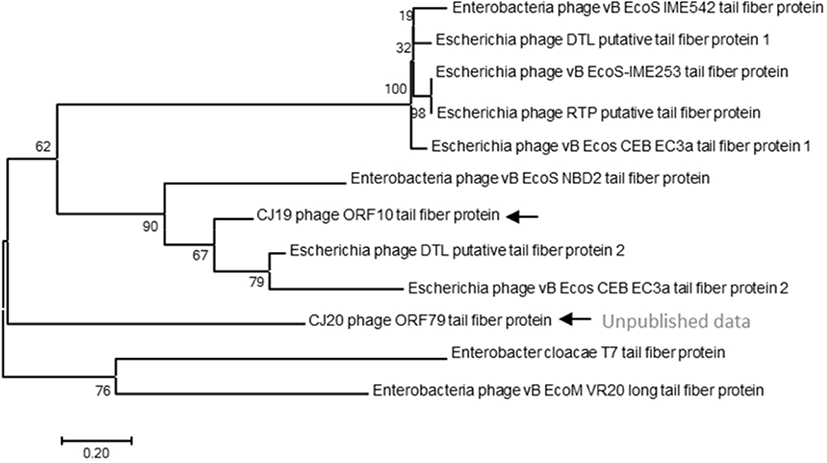
The evolutionary history of the phages was inferred using the neighbor-joining method, on based of the tail protein sequences of the isolated phage and 11 additional phages. The percentage of replicate trees in which the associated taxa clustered together in 1,000 bootstrap repetitions was shown next to the branches. The codon positions included were the first, second, and third positions and the noncoding sites. All positions containing gaps and missing data were eliminated. There were 764 positions in the final dataset. Evolutionary analyses were conducted using the MEGA 7 software (Penn State, University Park, PA, USA) (Mohammed and Cormican, 2015).
To determine the host range of phages ØCJ19 and ØCJ20, plaque assays were performed. The ØCJ20 (unpublished data) was introduced to discuss the genetic correlation of host-range and putative tail fiber proteins with phage ØCJ19. Each of the nine host strains were inoculated into 5 mL of 0.7% soft agar, which were poured onto agar plates. Then, 10 μL high-titer phage solution was spotted onto the top agar plate and incubated overnight at 37°C. After incubation, lysis zones were examined. The lytic activity of the phage was classified as clear (+) or no (–) reaction (Lee et al., 2016).
Results and Discussion
ØCJ19 morphology was observed by TEM. ØCJ19 consisted of an icosahedral head and a contractile tail, which appeared to have a sheath-like structure. Tail fibers were not observed, but according to the result of the annotation of ORFs, a tail fiber protein was present. ØCJ19 had an icosahedral capsid that was approximately 58 nm in diameter, and a tail of approximately 163 nm length and approximately 16 nm width. The sheath width increases approximately 6 nm during contraction (Fig. 1). The International Committee on Taxonomy of Viruses (ICTV) describes that the order Caudovirales consists of three families of tailed bacterial viruses: the families Myoviridae with contractile tails, Siphoviridae with long noncontractile tails, and Podoviridae with short noncontractile tails. The taxonomic structure of Myoviridae consists of six genera: T4-, P1-, P2-, Mu-, SP01-, and ØH-like viruses. Based on TEM observation, phage ØCJ19 belongs to the order Caudovirales, family Myoviridae, and genus T4-like viruses (Mayo and Haenni, 2006).
A microbial growth curve was drawn to determine the extent of the propagation of E. coli FC02 and the challenging time point of the phage in its exponential growth. The E. coli FC02 was grown at 37°C in LB broth and exhibited a lag phase of 1 h, an exponential phase of 1–7 h, and a stationary phase of 7–9 h after inoculation (Fig. 2A). The propagation of phages is known to be highly sensitive to external conditions (Wei and Zhao, 2018). Therefore, the optimal time point for challenge appeared to be 3 h in a shaking culture of E. coli FC02 at 37°C.
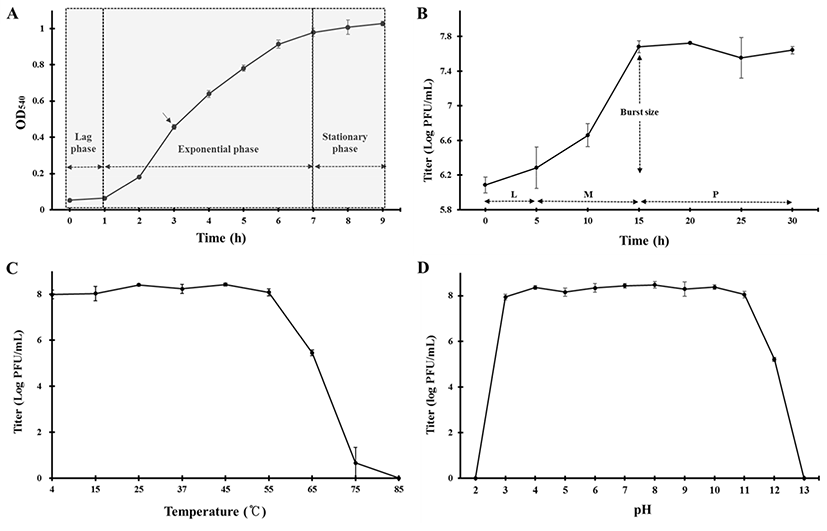
Fig. 2B shows the result of the one-step growth experiment of phage ØCJ19 at 37°C. This phage rapidly infects the host bacteria with a latent phase of 5 min or less, a maturation phase between 5 and 15 min, and an initial plateau phase at 15 min after infection. The burst size is approximately 20 phage particles/infected cell (Fig. 2B). In the case of phage vB_EcoS_HSE2, the latent phase and burst size are 30 min and 86 particles/infected cell, respectively (Peng and Yuan, 2018). The phage plays an important role as a predator of bacteria, and the rate at which bacteria are adsorbed is related to the density of the phages and the host bacteria (Abedon et al., 2003). It has been theorized that despite the smaller burst size, individual phages can grow rapidly by reducing the latent period (Abedon et al., 2001). A lower bacterial density has a longer latent period and a larger burst size. In other words, the lower bacterial densities are the most effectively exploited by producing more phage progeny per infected host cell. The longer the latent period, the longer it takes to acquire new phage progeny, but more phages can be produced. Here, the MOI of phage ØCJ19 had a host density 10 times higher than that of phage vB_EcoS_HSE2 (Peng and Yuan, 2018). Consequently, it is estimated that phage ØCJ19 has a shorter latent period and a smaller burst size than those of phage vB_EcoS_HSE2. Given that a phage has a long latent period, controlling host density and MOI value should be can considered to produce more particles.
ØCJ19 was exposed to various temperature and pH conditions. As shown in Fig. 2C, approximately 1.0×108 pfu/mL was stably maintained from 4°C to 55°C and began to reduce to 3.0×105 pfu/mL at 65°C; no plaque was observed at 85°C. The stability decreased dramatically at 75°C, and it can be concluded that the optimum temperature for stability was between 4°C and 55°C (Fig. 2C). Regarding the pH stability test, the phage was maintained at approximately 1.0×108 pfu/mL between pH 3 and 11, with no plaque observed below pH 2 or above pH 13. This result shows that phage ØCJ19 significantly loses its activity under strong acidic or basic conditions (Fig. 2D). As reported previously, the phage vB_EcoS_HSE2 of pathogenic E. coli was stable below 50°C and between pH 3 and 9 (Peng and Yuan, 2018). These results indicate that phage ØCJ19 is more stable at higher temperatures and a wider pH range than phage vB_EcoS_HSE2, even though phage ØCJ19 was exposed for 2 h compared to the 30 min exposure for phage vB_EcoS_HSE2 (Peng and Yuan, 2018).
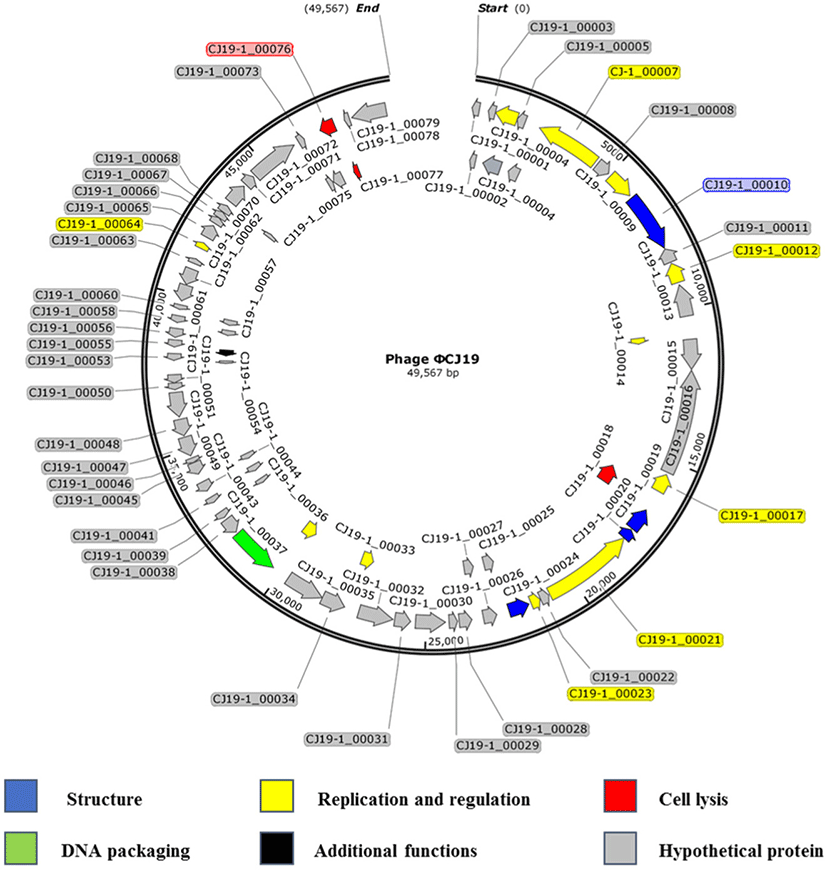
Sequencing analysis showed that the ØCJ19 genome was composed of 49,567 bases, and the GC content was 46.8%. A BLAST search of the GenBank database indicated that it was closely related to the Escherichia phage vB_EcoS_ESCO41 with 86.4% sequence identity (Fig. 3). The genome of ØCJ19 was shorter than that of vB_EcoS_ESCO41 by 1,233 bases. The annotation results showed that ØCJ19 had a total of 79 ORFs, including 4 genes responsible for the structure proteins, 11 for replication and regulation, 3 for cell lysis, and 1 for DNA packaging and additional functions (Table 1). Fifty-nine ORFs related to hypothetical proteins are not shown in Table 1. In particular, the BLAST analysis of the tail fiber protein annotated ORF 10 revealed that the T7 tail fiber protein showed 60.95% similarity to that of Enterobacter cloacae. There were no genes encoding lysogeny, toxins, virulence factors, or antibiotic resistance, suggesting that the phage can be used safely as a biological control agent. However, lytic phages that do not have toxic factors or antibiotic resistance genes require further study as biocontrol agents for ETEC strains. Sequencing analysis and annotated ORFs provide important information to establish the relationships between the phages in the database. However, it is challenging to evaluate the genetic biosafety of the phages through these analyses. Recently, NGS has become a powerful tool to support these analyses, but the functions of the encoded phage genes are still unknown (Romero-Calle et al., 2019). Therefore, functions of the unknown genes need to be studied further for safe use of phages.
A phylogenetic tree based on the DNA sequences encoding the tail fiber proteins of several phages was constructed. The gene sequences of the tail fiber proteins of 11 phages similar to the ØCJ19 phage were analyzed. Phylogenetic analysis used the MEGA7 program to perform 1,000 Bootstrapping to obtain 1,000 schematics. The tail fiber protein of phage ØCJ19 with annotated ORF 10 was phylogenetically the closest to the putative tail fiber protein of Escherichia phage DTL. The tail fiber proteins of phage ØCJ20 and phage ØCJ19 with the common host range were found to be phylogenetically relatively close (Fig. 4). Table 2 shows the result of the plaque assay revealing the susceptibility of ØCJ19 and ØCJ20 to E. coli strain infection. Out of nine host strains, ØCJ19 formed plaques against three strains, FC01, FC02, and BL21. Phage ØCJ20 infected five strains with a broad spectrum. Both phages commonly infected FC01, FC02, and BL21 as the host bacterium (Table 2).
| Strains tested2) (Escherichia coli) |
Susceptibility1) | |
|---|---|---|
| ØCJ192) | ØCJ202) | |
| FC01 | + | + |
| FC02 | + | + |
| FC03 | − | − |
| FC04 | − | + |
| FC05 | − | − |
| FC06 | − | − |
| FC07 | − | − |
| DH5α | − | + |
| BL21 | + | + |
The tail fiber protein genes of phage ØCJ19 and ØCJ20 were phylogenetically relatively close (Fig. 4). Taken together, the differences in the host range are possible because of the genetic variety in the genes corresponding to the tail fiber proteins. Similarly, the tail fiber gene of phage ØCJ20 is 2,219 bp, which is longer than the 1,920 bp of phage ØCJ19. In general, the specific adsorption of a phage to its host is determined by the tail fiber binding on the bacterial surface (Yosef et al., 2017). As a feasible approach to change or expand the host range of the phage, transduction and mutation of the tail fiber gene have been studied because tail fibers are known to have proteins that recognize bacterial surfaces specifically during viral infection stages (Yoichi et al., 2005). It is assumed that the host-range differences between ØCJ19 and ØCJ20 are owing to the genetic differences in the tail fiber genes. It is necessary to further study the relationship between the genomic size of phages and their host range spectrum.
Previous studies have demonstrated the potential role of phage in controlling pathogenic E. coli in animal foods (Lee et al., 2017; Yang et al., 2017). These phages were reported to exhibit lytic activity against ETEC strains causing porcine post-weaning diarrhea (Jamalludeen et al., 2007). In addition, it has been reported that lytic coliphage JS09 isolated from sewage in China pig farms infects antibiotic-resistant avian pathogenic E. coli and ETEC (Zhou et al., 2015). As a result of mixing three lytic phages with a cocktail and applying them to rifampin-resistant E. coli O157:H7 contaminated beef surfaces, 7 out of 9 beef samples showed no E. coli O157:H7 contamination (O’Flynn et al., 2004). These results indicate that applying phage to food surfaces is a reasonable approach to food preservation and can be applied to other meats.
Conclusions
In this study, phage ØCJ19 was evaluated to safety and stability for application of phage to ETEC-contaminated foods of animal origin, and proteins related to toxicity were analyzed through genetic analysis. Phage ØCJ19 has high stability, and the results of gene annotation showed that the phage contained no toxic genes. This result showed the potential to reduce the contamination of meat and improve food safety by applying phage ØCJ19 to foods of animal origin and could be the basis for establishing a safety management system for foods of animal origin.













